Posts Tagged Analytics
Building Data Lakes on AWS with Kafka Connect, Debezium, Apicurio Registry, and Apache Hudi
Posted by Gary A. Stafford in Analytics, AWS, Cloud on February 28, 2023
Learn how to build a near real-time transactional data lake on AWS using a combination of Open Source Software (OSS) and AWS Services
Introduction
In the following post, we will explore one possible architecture for building a near real-time transactional data lake on AWS. The data lake will be built using a combination of open source software (OSS) and fully-managed AWS services. Red Hat’s Debezium, Apache Kafka, and Kafka Connect will be used for change data capture (CDC). In addition, Apache Spark, Apache Hudi, and Hudi’s DeltaStreamer will be used to manage the data lake. To complete our architecture, we will use several fully-managed AWS services, including Amazon RDS, Amazon MKS, Amazon EKS, AWS Glue, and Amazon EMR.

Source Code
The source code, configuration files, and a list of commands shown in this post are open-sourced and available on GitHub.
Kafka
According to the Apache Kafka documentation, “Apache Kafka is an open-source distributed event streaming platform used by thousands of companies for high-performance data pipelines, streaming analytics, data integration, and mission-critical applications.” For this post, we will use Apache Kafka as the core of our change data capture (CDC) process. According to Wikipedia, “change data capture (CDC) is a set of software design patterns used to determine and track the data that has changed so that action can be taken using the changed data. CDC is an approach to data integration that is based on the identification, capture, and delivery of the changes made to enterprise data sources.” We will discuss CDC in greater detail later in the post.
There are several options for Apache Kafka on AWS. We will use AWS’s fully-managed Amazon Managed Streaming for Apache Kafka (Amazon MSK) service. Alternatively, you could choose industry-leading SaaS providers, such as Confluent, Aiven, Redpanda, or Instaclustr. Lastly, you could choose to self-manage Apache Kafka on Amazon Elastic Compute Cloud (Amazon EC2) or Amazon Elastic Kubernetes Service (Amazon EKS).
Kafka Connect
According to the Apache Kafka documentation, “Kafka Connect is a tool for scalably and reliably streaming data between Apache Kafka and other systems. It makes it simple to quickly define connectors that move large collections of data into and out of Kafka. Kafka Connect can ingest entire databases or collect metrics from all your application servers into Kafka topics, making the data available for stream processing with low latency.”
There are multiple options for Kafka Connect on AWS. You can use AWS’s fully-managed, serverless Amazon MSK Connect. Alternatively, you could choose a SaaS provider or self-manage Kafka Connect yourself on Amazon Elastic Compute Cloud (Amazon EC2) or Amazon Elastic Kubernetes Service (Amazon EKS). I am not a huge fan of Amazon MSK Connect during development. In my opinion, iterating on the configuration for a new source and sink connector, especially with transforms and external registry dependencies, can be painfully slow and time-consuming with MSK Connect. I find it much faster to develop and fine-tune my sink and source connectors using a self-managed version of Kafka Connect. For Production workloads, you can easily port the configuration from the native Kafka Connect connector to MSK Connect. I am using a self-managed version of Kafka Connect for this post, running in Amazon EKS.

Debezium
According to the Debezium documentation, “Debezium is an open source distributed platform for change data capture. Start it up, point it at your databases, and your apps can start responding to all of the inserts, updates, and deletes that other apps commit to your databases.” Regarding Kafka Connect, according to the Debezium documentation, “Debezium is built on top of Apache Kafka and provides a set of Kafka Connect compatible connectors. Each of the connectors works with a specific database management system (DBMS). Connectors record the history of data changes in the DBMS by detecting changes as they occur and streaming a record of each change event to a Kafka topic. Consuming applications can then read the resulting event records from the Kafka topic.”
Source Connectors
We will use Kafka Connect along with three Debezium connectors, MySQL, PostgreSQL, and SQL Server, to connect to three corresponding Amazon Relational Database Service (RDS) databases and perform CDC. Changes from the three databases will be represented as messages in separate Kafka topics. In Kafka Connect terminology, these are referred to as Source Connectors. According to Confluent.io, a leader in the Kafka community, “source connectors ingest entire databases and stream table updates to Kafka topics.”
Sink Connector
We will stream the data from the Kafka topics into Amazon S3 using a sink connector. Again, according to Confluent.io, “sink connectors deliver data from Kafka topics to secondary indexes, such as Elasticsearch, or batch systems such as Hadoop for offline analysis.” We will use Confluent’s Amazon S3 Sink Connector for Confluent Platform. We can use Confluent’s sink connector without depending on the entire Confluent platform.
There is also an option to use the Hudi Sink Connector for Kafka, which promises to greatly simplify the processes described in this post. However, the RFC for this Hudi feature appears to be stalled. Last updated in August 2021, the RFC is still in the initial “Under Discussion” phase. Therefore, I would not recommend the connectors use in Production until it is GA (General Availability) and gains broader community support.
Securing Database Credentials
Whether using Amazon MSK Connect or self-managed Kafka Connect, you should ensure your database, Kafka, and schema registry credentials, and other sensitive configuration values are secured. Both MSK Connect and self-managed Kafka Connect can integrate with configuration providers that implement the ConfigProvider class interface, such as AWS Secrets Manager, HashiCorp Vault, Microsoft Azure Key Vault, and Google Cloud Secrets Manager.
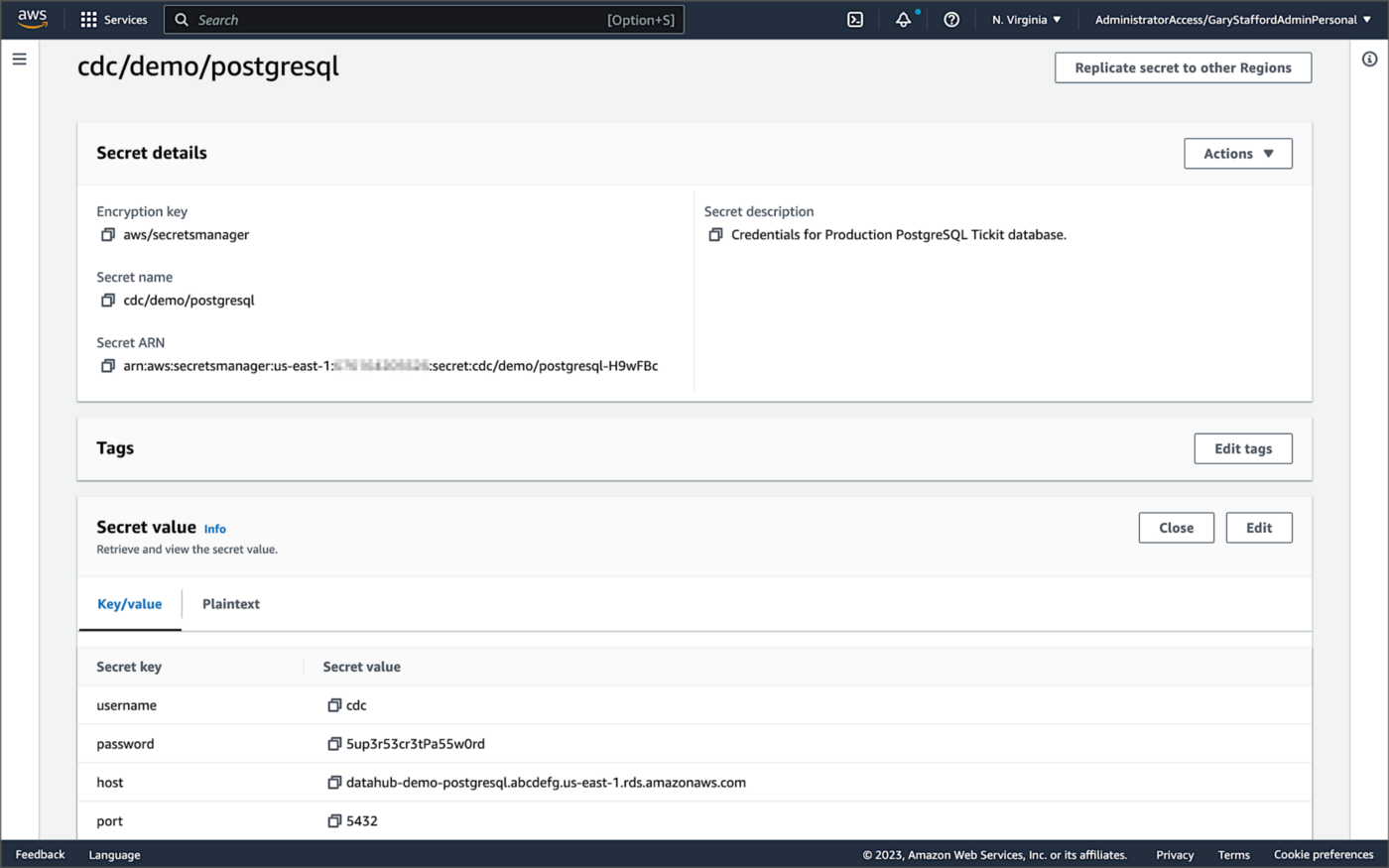
For self-managed Kafka Connect, I prefer Jeremy Custenborder’s kafka-config-provider-aws plugin. This plugin provides integration with AWS Secrets Manager. A complete list of Jeremy’s providers can be found on GitHub. Below is a snippet of the Secrets Manager configuration from the connect-distributed.properties files, which is read by Apache Kafka Connect at startup.
https://garystafford.medium.com/media/db6ff589c946fcd014546c56d0255fe5
Apache Avro
The message in the Kafka topic and corresponding objects in Amazon S3 will be stored in Apache Avro format by the CDC process. The Apache Avro documentation states, “Apache Avro is the leading serialization format for record data, and the first choice for streaming data pipelines.” Avro provides rich data structures and a compact, fast, binary data format.
Again, according to the Apache Avro documentation, “Avro relies on schemas. When Avro data is read, the schema used when writing it is always present. This permits each datum to be written with no per-value overheads, making serialization both fast and small. This also facilitates use with dynamic, scripting languages, since data, together with its schema, is fully self-describing.” When Avro data is stored in a file, its schema can be stored with it so any program may process files later.
Alternatively, the schema can be stored separately in a schema registry. According to Apicurio Registry, “in the messaging and event streaming world, data that are published to topics and queues often must be serialized or validated using a Schema (e.g. Apache Avro, JSON Schema, or Google protocol buffers). Schemas can be packaged in each application, but it is often a better architectural pattern to instead register them in an external system [schema registry] and then referenced from each application.”
Schema Registry
Several leading open-source and commercial schema registries exist, including Confluent Schema Registry, AWS Glue Schema Registry, and Red Hat’s open-source Apicurio Registry. In this post, we use a self-managed version of Apicurio Registry running on Amazon EKS. You can relatively easily substitute AWS Glue Schema Registry if you prefer a fully-managed AWS service.
Apicurio Registry
According to the documentation, Apicurio Registry supports adding, removing, and updating OpenAPI, AsyncAPI, GraphQL, Apache Avro, Google protocol buffers, JSON Schema, Kafka Connect schema, WSDL, and XML Schema (XSD) artifact types. Furthermore, content evolution can be controlled by enabling content rules, including validity and compatibility. Lastly, the registry can be configured to store data in various backend storage systems depending on the use case, including Kafka (e.g., Amazon MSK), PostgreSQL (e.g., Amazon RDS), and Infinispan (embedded).

Data Lake Table Formats
Three leading open-source, transactional data lake storage frameworks enable building data lake and data lakehouse architectures: Apache Iceberg, Linux Foundation Delta Lake, and Apache Hudi. They offer comparable features, such as ACID-compliant transactional guarantees, time travel, rollback, and schema evolution. Using any of these three data lake table formats will allow us to store and perform analytics on the latest version of the data in the data lake originating from the three Amazon RDS databases.
Apache Hudi
According to the Apache Hudi documentation, “Apache Hudi is a transactional data lake platform that brings database and data warehouse capabilities to the data lake.” The specifics of how the data is laid out as files in your data lake depends on the Hudi table type you choose, either Copy on Write (CoW) or Merge On Read (MoR).
Like Apache Iceberg and Delta Lake, Apache Hudi is partially supported by AWS’s Analytics services, including AWS Glue, Amazon EMR, Amazon Athena, Amazon Redshift Spectrum, and AWS Lake Formation. In general, CoW has broader support on AWS than MoR. It is important to understand the limitations of Apache Hudi with each AWS analytics service before choosing a table format.
If you are looking for a fully-managed Cloud data lake service built on Hudi, I recommend Onehouse. Born from the roots of Apache Hudi and founded by its original creator, Vinoth Chandar (PMC chair of the Apache Hudi project), the Onehouse product and its services leverage OSS Hudi to offer a data lake platform similar to what companies like Uber have built.
Hudi DeltaStreamer
Hudi also offers DeltaStreamer (aka HoodieDeltaStreamer) for streaming ingestion of CDC data. DeltaStreamer can be run once or continuously, using Apache Spark, similar to an Apache Spark Structured Streaming job, to capture changes to the raw CDC data and write that data to a different part of our data lake.
DeltaStreamer also works with Amazon S3 Event Notifications instead of running continuously. According to DeltaStreamer’s documentation, Amazon S3 object storage provides an event notification service to post notifications when certain events happen in your S3 bucket. AWS will put these events in Amazon Simple Queue Service (Amazon SQS). Apache Hudi provides an S3EventsSource that can read from Amazon SQS to trigger and process new or changed data as soon as it is available on Amazon S3.
Sample Data for the Data Lake
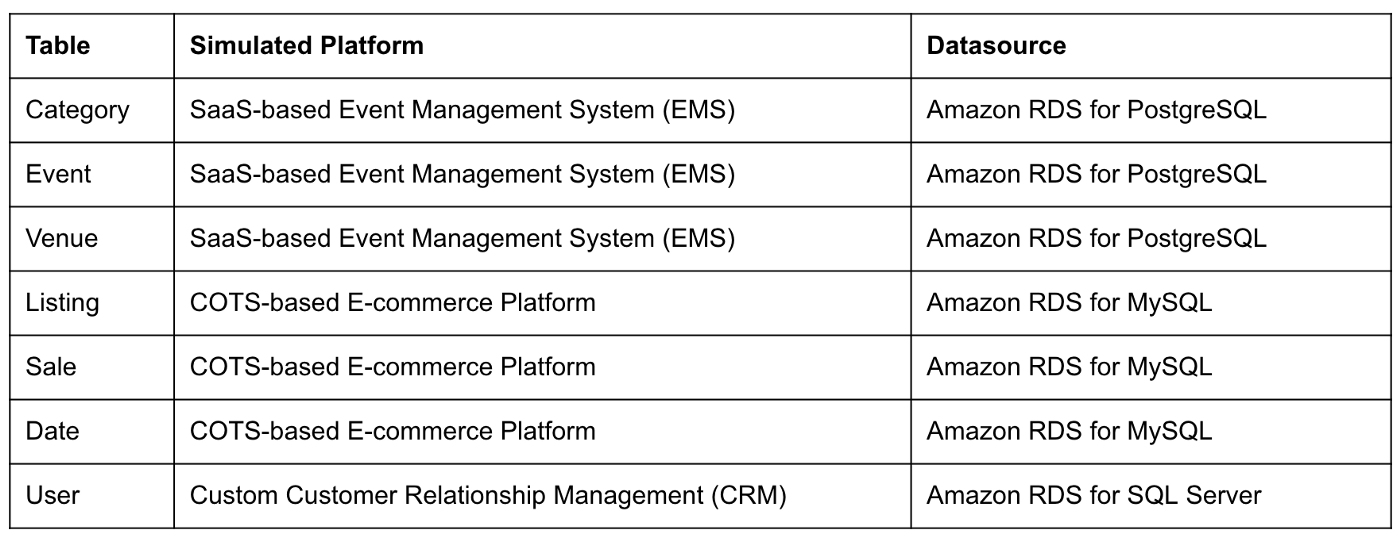
The data used in this post is from the TICKIT sample database. The TICKIT database represents the backend data store for a platform that brings buyers and sellers of tickets to entertainment events together. It was initially designed to demonstrate Amazon Redshift. The TICKIT database is a small database with approximately 425K rows of data across seven tables: Category, Event, Venue, User, Listing, Sale, and Date.
A data lake most often contains data from multiple sources, each with different storage formats, protocols, and connection methods. To simulate these data sources, I have separated the TICKIT database tables to represent three typical enterprise systems, including a commercial off-the-shelf (COTS) E-commerce platform, a custom Customer Relationship Management (CRM) platform, and a SaaS-based Event Management System (EMS). Each simulated system uses a different Amazon RDS database engine, including MySQL, PostgreSQL, and SQL Server.
Enabling CDC for Amazon RDS
To use Debezium for CDC with Amazon RDS databases, minor changes to the default database configuration for each database engine are required.
CDC for PostgreSQL
Debezium has detailed instructions regarding configuring CDC for Amazon RDS for PostgreSQL. Database parameters specify how the database is configured. According to the Debezium documentation, for PostgreSQL, set the instance parameter rds.logical_replication to 1 and verify that the wal_level parameter is set to logical. It is automatically changed when the rds.logical_replication parameter is set to 1. This parameter is adjusted using an Amazon RDS custom parameter group. According to the AWS documentation, “with Amazon RDS, you manage database configuration by associating your DB instances and Multi-AZ DB clusters with parameter groups. Amazon RDS defines parameter groups with default settings. You can also define your own parameter groups with customized settings.”

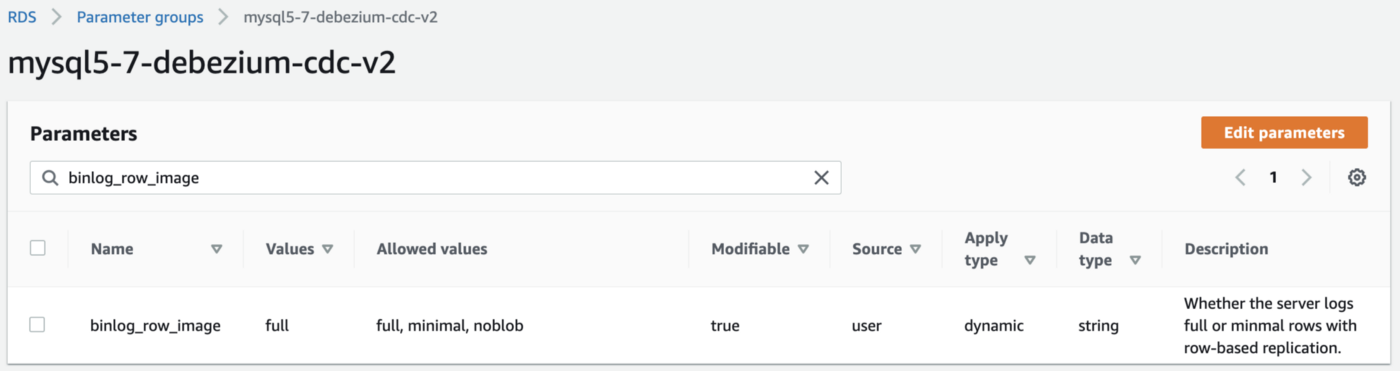
CDC for MySQL
Similarly, Debezium has detailed instructions regarding configuring CDC for MySQL. Like PostgreSQL, MySQL requires a custom DB parameter group.


CDC for SQL Server
Lastly, Debezium has detailed instructions for configuring CDC with Microsoft SQL Server. Enabling CDC requires enabling CDC on the SQL Server database and table(s) and creating a new filegroup and associated file. Debezium recommends locating change tables in a different filegroup than you use for source tables. CDC is only supported with Microsoft SQL Server Standard Edition and higher; Express and Web Editions are not supported.
Kafka Connect Source Connectors
There is a Kafka Connect source connector for each of the three Amazon RDS databases, all of which use Debezium. CDC is performed, moving changes from the three databases into separate Kafka topics in Apache Avro format using the source connectors. The connector configuration is nearly identical whether you are using Amazon MSK Connect or a self-managed version of Kafka Connect.
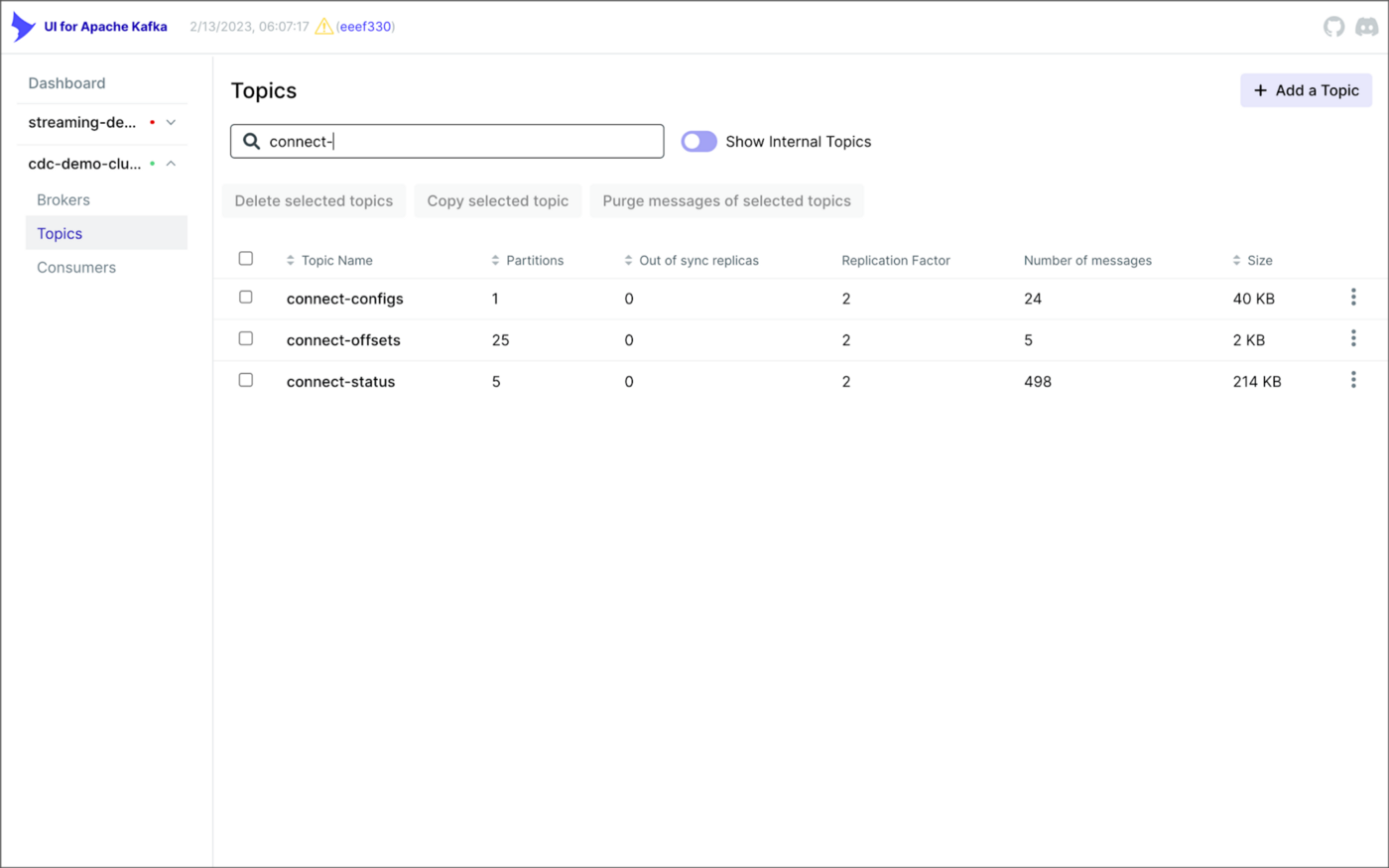
As shown above, I am using the UI for Apache Kafka by Provectus, self-managed on Amazon EKS, in this post.
PostgreSQL Source Connector
The source_connector_postgres_kafka_avro_tickit source connector captures all changes to the three tables in the Amazon RDS for PostgreSQL ticket database’s ems schema: category, event, and venue. These changes are written to three corresponding Kafka topics as Avro-format messages: tickit.ems.category, tickit.ems.event, and tickit.ems.venue. The messages are transformed by the connector using Debezium’s unwrap transform. Schemas for the messages are written to Apicurio Registry.
MySQL Source Connector
The source_connector_mysql_kafka_avro_tickit source connector captures all changes to the three tables in the Amazon RDS for PostgreSQL ecomm database: date, listing, and sale. These changes are written to three corresponding Kafka topics as Avro-format messages: tickit.ecomm.date, tickit.ecomm.listing, and tickit.ecomm.sale.
SQL Server Source Connector
Lastly, the source_connector_mssql_kafka_avro_tickit source connector captures all changes to the single user table in the Amazon RDS for SQL Server ticket database’s crm schema. These changes are written to a corresponding Kafka topic as Avro-format messages: tickit.crm.user.
If using a self-managed version of Kafka Connect, we can deploy, manage, and monitor the source and sink connectors using Kafka Connect’s RESTful API (Connect API).

Once the three source connectors are running, we should see seven new Kafka topics corresponding to the seven TICKIT database tables from the three Amazon RDS database sources. There will be approximately 425K messages consuming 147 MB of space in Avro’s binary format.
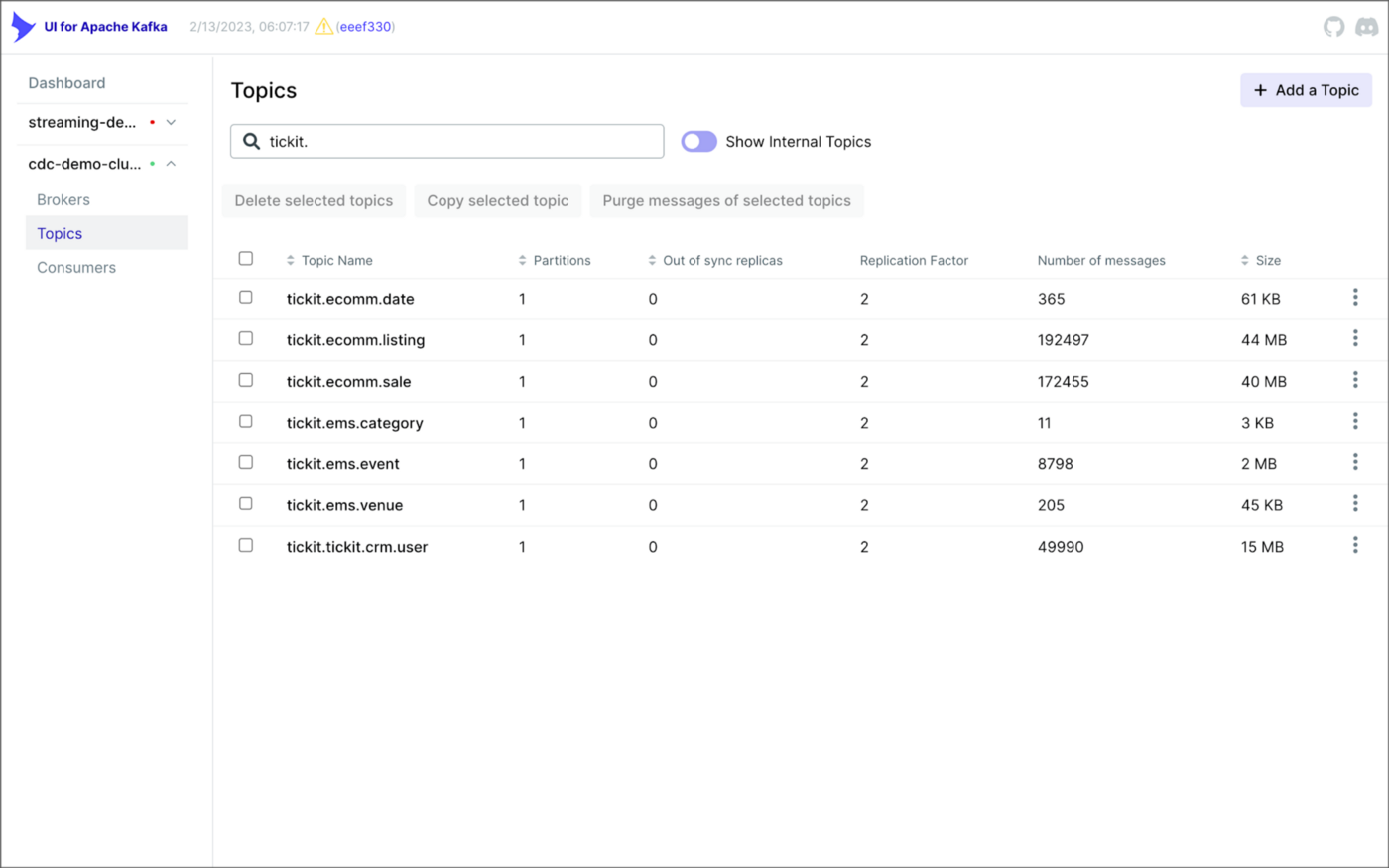
Avro’s binary format and the use of a separate schema ensure the messages consume minimal Kafka storage space. For example, the average message Value (payload) size in the tickit.ecomm.sale topic is a minuscule 98 Bytes. Resource management is critical when you are dealing with hundreds of millions or often billions of daily Kafka messages as a result of the CDC process.

From within the Apicurio Registry UI, we should now see Value schema artifacts for each of the seven Kafka topics.
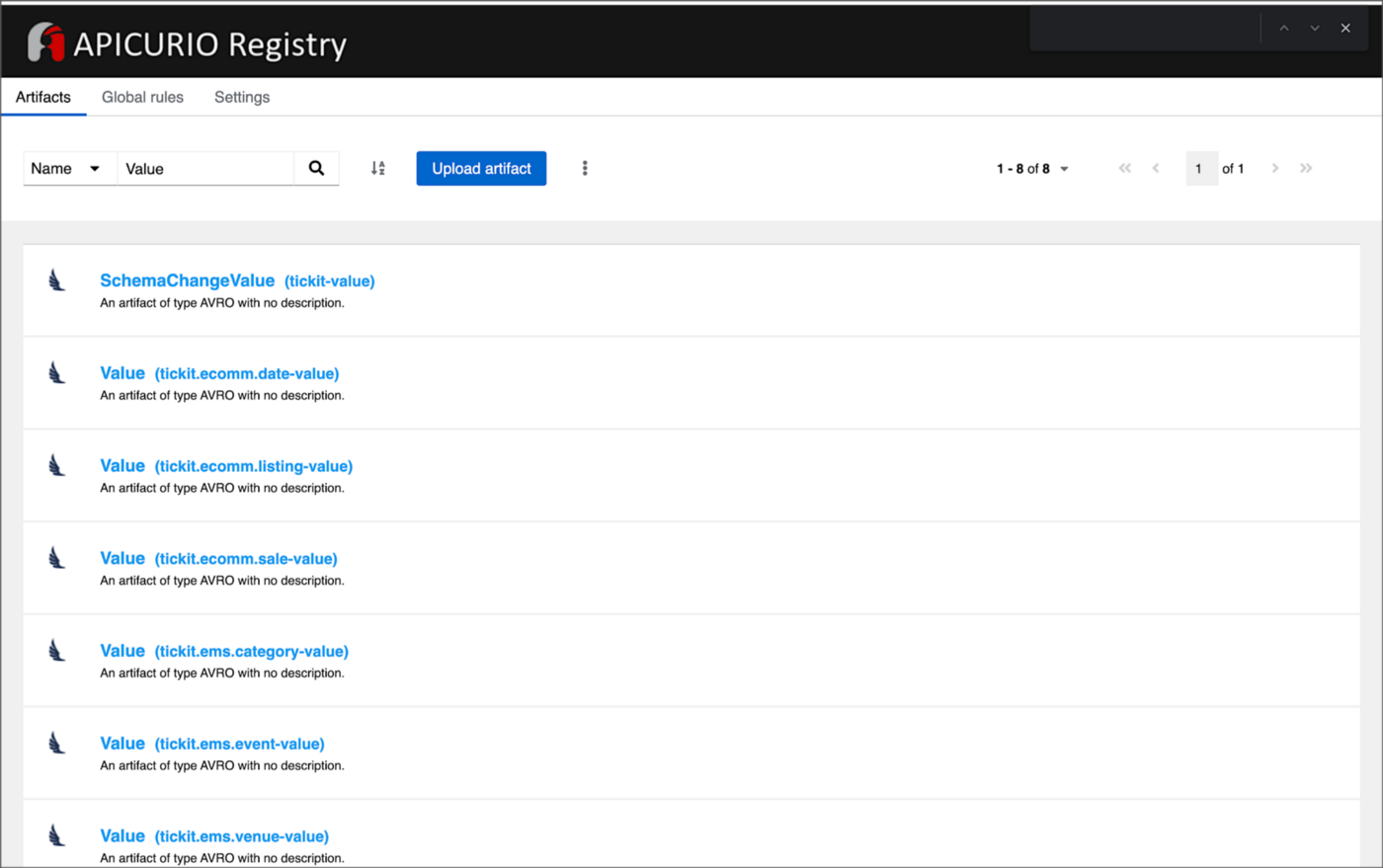
Also from within the Apicurio Registry UI, we can examine details of individual Value schema artifacts.

Kafka Connect Sink Connector
In addition to the three source connectors, there is a single Kafka Connect sink connector, sink_connector_kafka_s3_avro_tickit. The sink connector copies messages from the seven Kafka topics, all prefixed with tickit, to the Bronze area of our Amazon S3-based data lake.

The connector uses Confluent’s S3 Sink Connector (S3SinkConnector). Like Kafka, the connector writes all changes to Amazon S3 in Apache Avro format, with the schemas already stored in Apicurio Registry.
Once the sink connector and the three source connectors are running with Kafka Connect, we should see a series of seven subdirectories within the Bronze area of the data lake.

Confluent offers a choice of data partitioning strategies. The sink connector we have deployed uses the Daily Partitioner. According to the documentation, the io.confluent.connect.storage.partitioner.DailyPartitioner is equivalent to the TimeBasedPartitioner with path.format='year'=YYYY/'month'=MM/'day'=dd and partition.duration.ms=86400000 (one day for one S3 object in each daily directory). Below is an example of Avro files (S3 objects) containing changes to the sale table. The objects are partitioned by the year, month, and day they were written to the S3 bucket.

Message Transformation
Previously, while discussing the Kafka Connect source connectors, I mentioned that the connector transforms the messages using the unwrap transform. By default, the changes picked up by Debezium during the CDC process contain several additional Debezium-related fields of data. An example of an untransformed message generated by Debezium from the PostgreSQL sale table is shown below.
When the PostgreSQL connector first connects to a particular PostgreSQL database, it starts by performing a consistent snapshot of each database schema. All existing records are captured. Unlike an UPDATE ("op" : "u") or a DELETE ("op" : "d"), these initial snapshot messages, like the one shown below, represent a READ operation ("op" : "r"). As a result, there is no before data only after.
According to Debezium’s documentation, “Debezium provides a single message transformation [SMT] that crosses the bridge between the complex and simple formats, the UnwrapFromEnvelope SMT.” Below, we see the results of Debezium’s unwrap transform of the same message. The unwrap transform flattens the nested JSON structure of the original message, adds the __deleted field, and includes fields based on the list included in the source connector’s configuration. These fields are prefixed with a double underscore (e.g., __table).
In the next section, we examine Apache Hudi will manage the data lake. An example of the same message, managed with Hudi, is shown below. Note the five additional Hudi data fields, prefixed with _hoodie_.
Apache Hudi
With database changes flowing into the Bronze area of our data lake, we are ready to use Apache Hudi to provide data lake capabilities such as ACID transactional guarantees, time travel, rollback, and schema evolution. Using Hudi allows us to store and perform analytics on a specific time-bound version of the data in the data lake. Without Hudi, a query for a single row of data could return multiple results, such as an initial CREATE record, multiple UPDATE records, and a DELETE record.
DeltaStreamer
Using Hudi’s DeltaStreamer, we will continuously stream changes from the Bronze area of the data lake to a Silver area, which Hudi manages. We will run DeltaStreamer continuously, similar to an Apache Spark Structured Streaming job, using Apache Spark on Amazon EMR. Running DeltaStreamer requires using a spark-submit command that references a series of configuration files. There is a common base set of configuration items and a group of configuration items specific to each database table. Below, we see the base configuration, base.properties.
The base configuration is referenced by each of the table-specific configuration files. For example, below, we see the tickit.ecomm.sale.properties configuration file for DeltaStreamer.
To run DeltaStreamer, you can submit an EMR Step or use EMR’s master node to run a spark-submit command on the cluster. Below, we see an example of the DeltaStreamer spark-submit command using the Merge on Read (MoR) Hudi table type.
Next, we see the same example of the DeltaStreamer spark-submit command using the Copy on Write (CoW) Hudi table type.
For this post’s demonstration, we will run a single-table DeltaStreamer Spark job, with one process for each table using CoW. Alternately, Hudi offers HoodieMultiTableDeltaStreamer, a wrapper on top of HoodieDeltaStreamer, which enables the ingestion of multiple tables at a single time into Hudi datasets. Currently, HoodieMultiTableDeltaStreamer only supports sequential processing of tables to be ingested and COPY_ON_WRITE storage type.
Below, we see an example of three DeltaStreamer Spark jobs running continuously for the date, listing, and sale tables.

Once DeltaStreamer is up and running, we should see a series of subdirectories within the Silver area of the data lake, all managed by Apache Hudi.
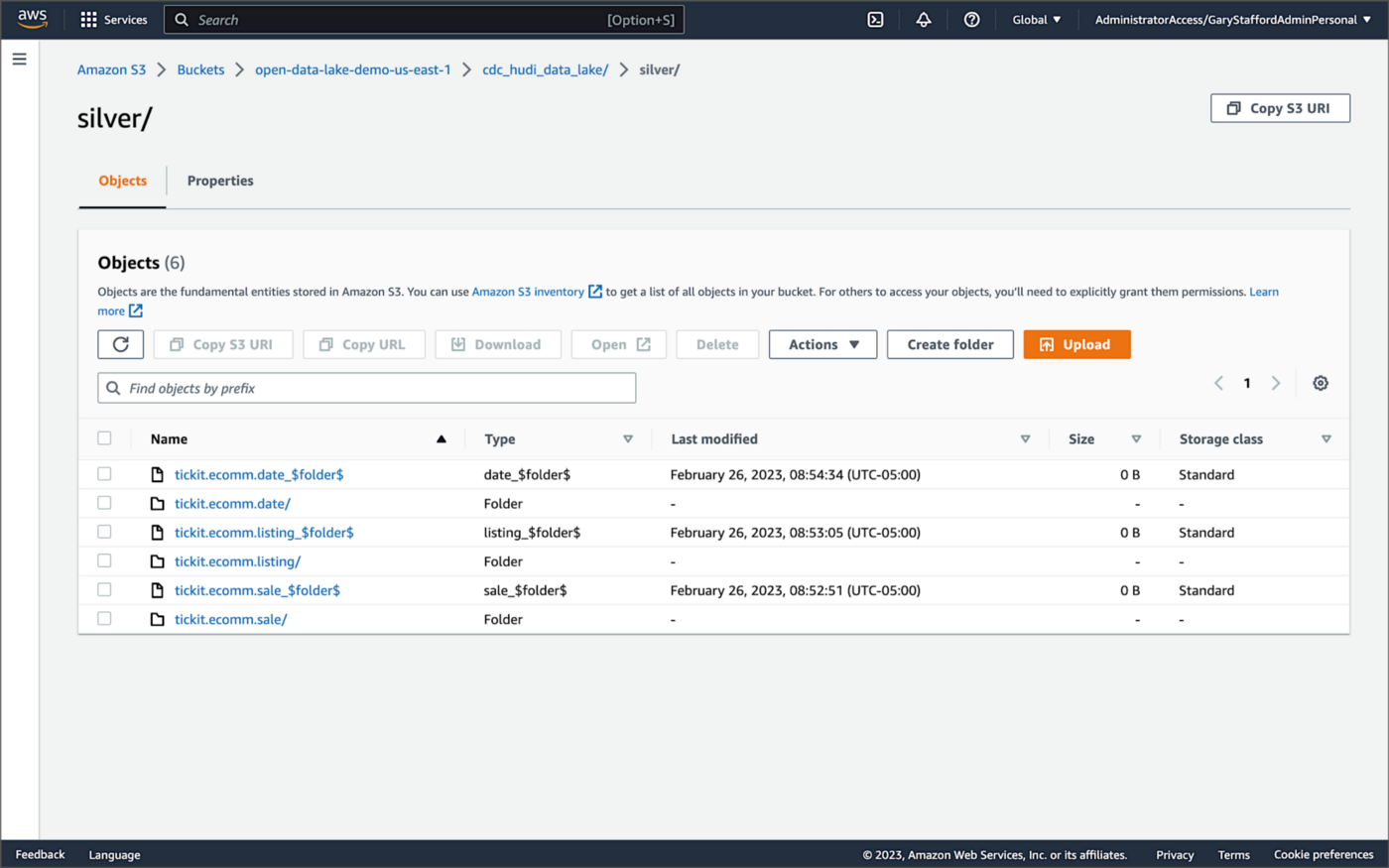
Within each subdirectory, partitioned by the table name, is a series of Apache Parquet files, along with other Apache Hudi-specific files. The specific folder structure and files depend on MoR or Cow.

AWS Glue Data Catalog
For this post, we are using an AWS Glue Data Catalog, an Apache Hive-compatible metastore, to persist technical metadata stored in the Silver area of the data lake, managed by Apache Hudi. The AWS Glue Data Catalog database, tickit_cdc_hudi, will be automatically created the first time DeltaStreamer runs.
Using DeltaStreamer with the table type of MERGE_ON_READ, there would be two tables in the AWS Glue Data Catalog database for each original table. According to Amazon’s EMR documentation, “Merge on Read (MoR) — Data is stored using a combination of columnar (Parquet) and row-based (Avro) formats. Updates are logged to row-based delta files and are compacted as needed to create new versions of the columnar files.” Hudi creates two tables in the Hive metastore for MoR, a table with the name that you specified, which is a read-optimized view (appended with _ro), and a table with the same name appended with _rt, which is a real-time view. You can query both tables.
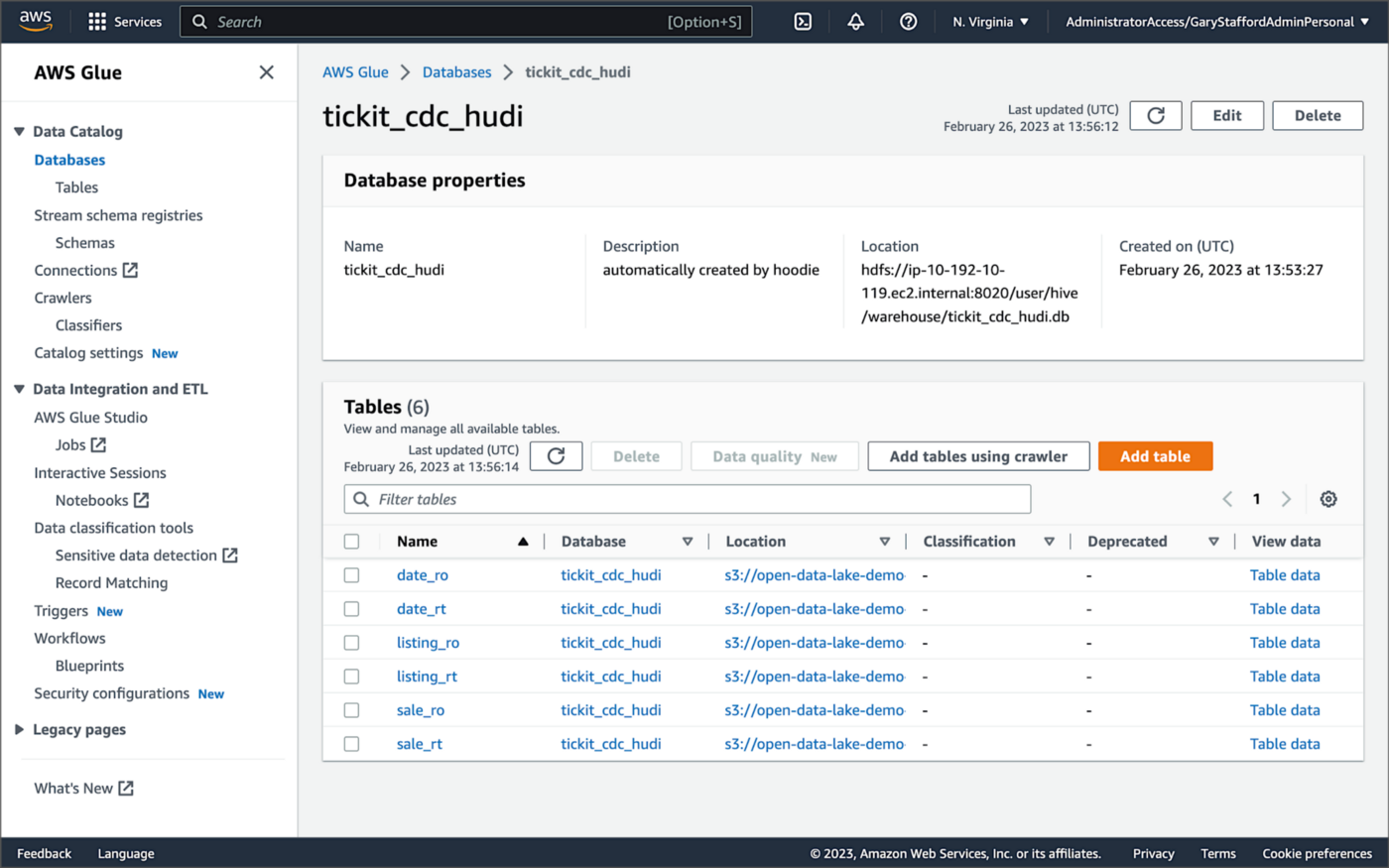
According to Amazon’s EMR documentation, “Copy on Write (CoW) — Data is stored in a columnar format (Parquet), and each update creates a new version of files during a write. CoW is the default storage type.” Using COPY_ON_WRITE with DeltaStreamer, there is only a single Hudi table in the AWS Glue Data Catalog database for each corresponding database table.
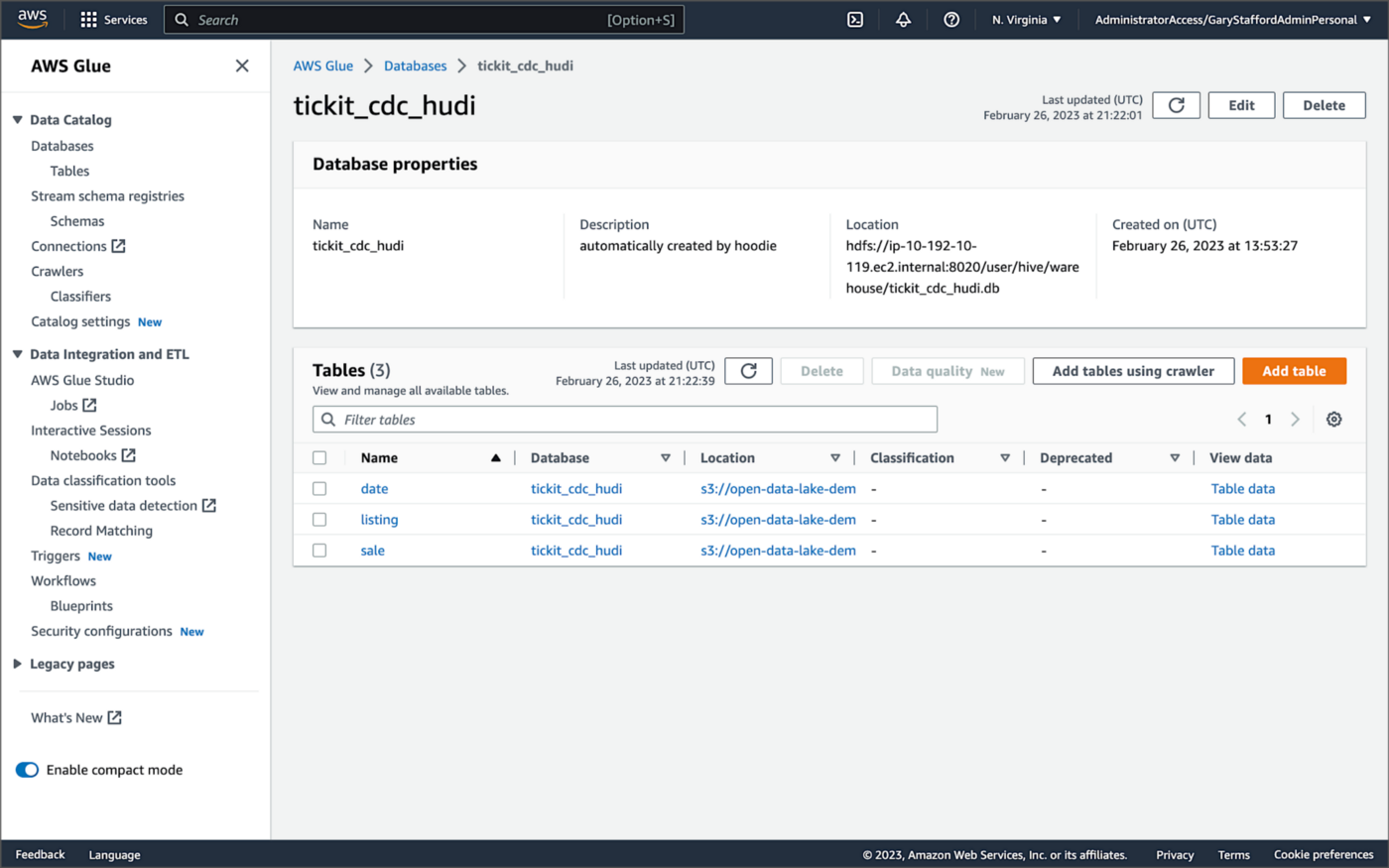
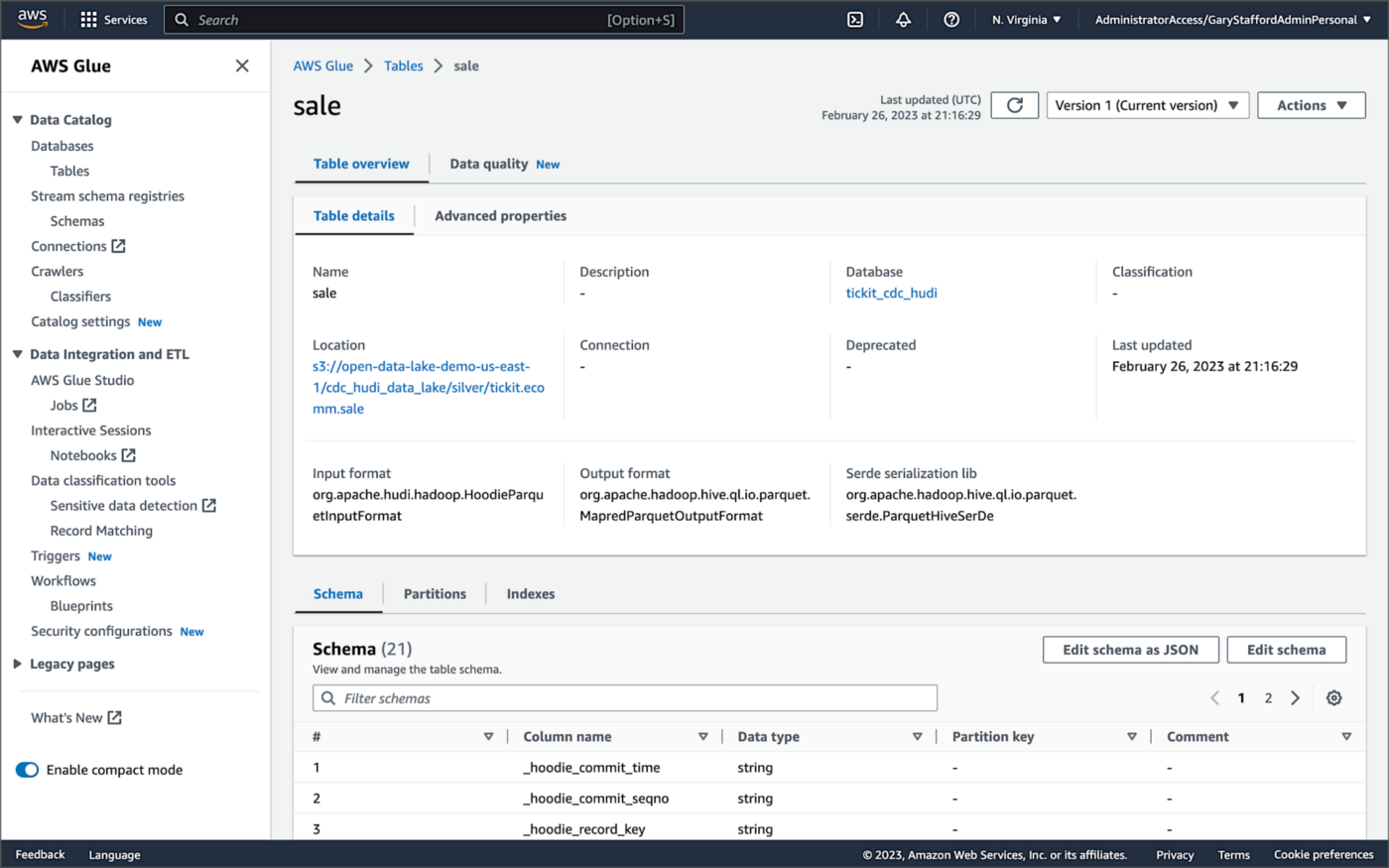
Examining an individual table in the AWS Glue Data Catalog database, we can see details like the table schema, location of underlying data in S3, input/output formats, Serde serialization library, and table partitions.
Database Changes and Data Lake
We are using Kafka Connect, Apache Hudi, and Hudi’s DeltaStreamer to maintain data parity between our databases and the data lake. Let’s look at an example of how a simple data change is propagated from a database to Kafka, then to the Bronze area of the data lake, and finally to the Hudi-managed Silver area of the data lake, written using the CoW table format.
First, we will make a simple update to a single record in the MySQL database’s sale table with the salesid = 200.
Almost immediately, we can see the change picked up by the Kafka Connect Debezium MySQL source connector.

Almost immediately, in the Bronze area of the data lake, we can see we have a new Avro-formatted file (S3 object) containing the updated record in the partitioned sale subdirectory.

If we examine the new object in the Bronze area of the data lake, we will see a row representing the updated record with the salesid = 200. Note how the operation is now an UPDATE versus a READ ("op" : "u").
Next, in the corresponding Silver area of the data lake, managed by Hudi, we should also see a new Parquet file that contains a record of the change. In this example, the file was written approximately 26 seconds after the original change to the database. This is the end-to-end time from database change to when the updated data is queryable in the data lake.
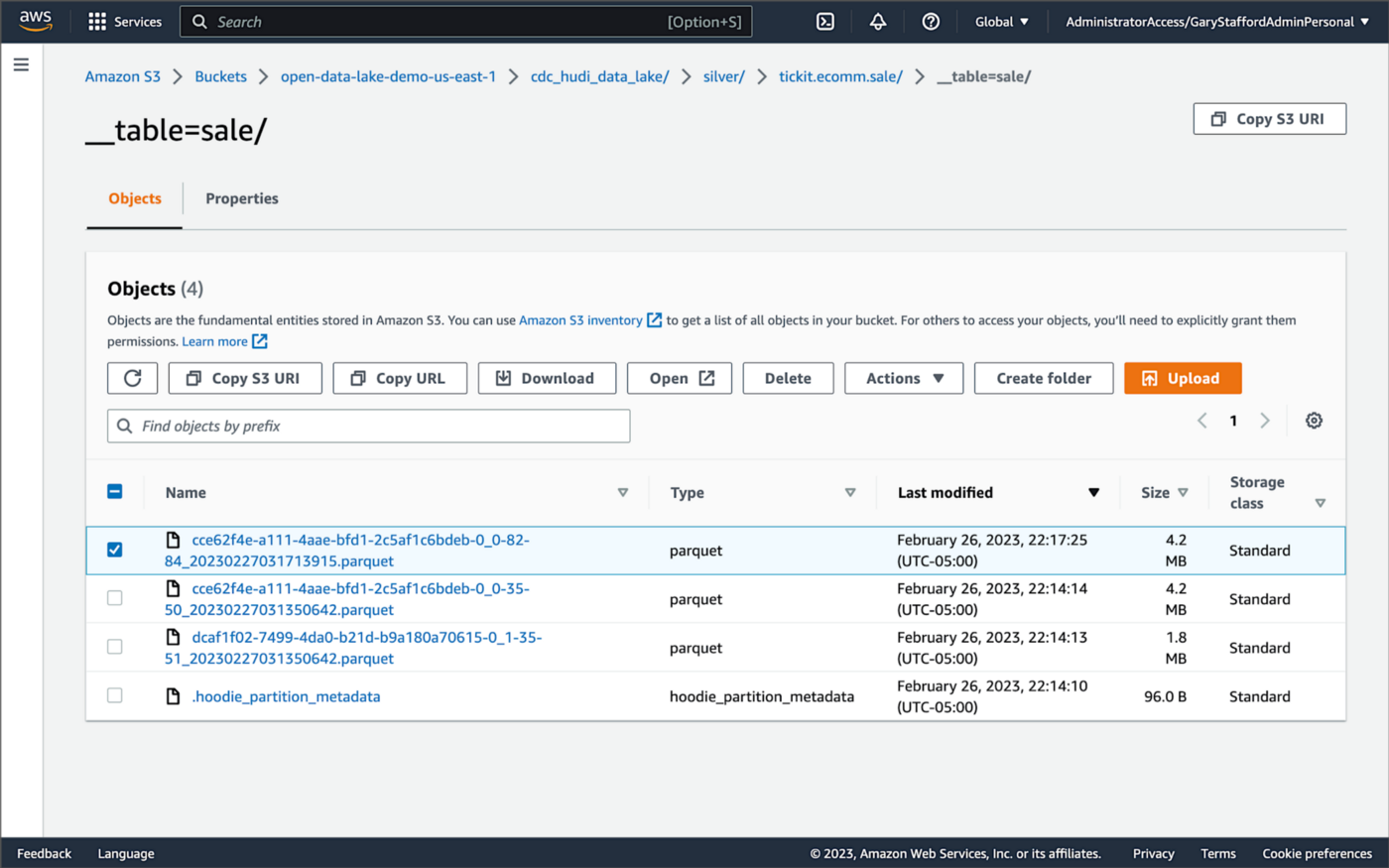
Similarly, if we examine the new object in the Silver area of the data lake, we will see a row representing the updated record with the salesid = 200. The record was committed approximately 15 seconds after the original database change.
Querying Hudi Tables with Amazon EMR
Using an EMR Notebook, we can query the updated database record stored in Amazon S3 using Hudi’s CoW table format. First, running a simple Spark SQL query for the record with salesid = 200, returns the latest record, reflecting the changes as indicated by the UPDATE operation value (u) and the _hoodie_commit_time = 2023-02-27 03:17:13.915 UTC.

Hudi Time Travel Query
We can also run a Hudi Time Travel Query, with an as.of.instance set to some arbitrary point in the future. Again, the latest record is returned, reflecting the changes as indicated by the UPDATE operation value (u) and the _hoodie_commit_time = 2023-02-27 03:17:13.915 UTC.

We can run the same Hudi Time Travel Query with an as.of.instance set to on or after the original records were written by DeltaStreamer (_hoodie_commit_time = 2023-02-27 03:13:50.642), but some arbitrary time before the updated record was written (_hoodie_commit_time = 2023-02-27 03:17:13.915). This time, the original record is returned as indicated by the READ operation value (r) and the _hoodie_commit_time = 2023-02-27 03:13:50.642. This is one of the strengths of Apache Hudi, the ability to query data in the present or at any point in the past.

Hudi Point in Time Query
We can also run a Hudi Point in Time Query, with a time range set from the beginning of time until some arbitrary point in the future. Again, the latest record is returned, reflecting the changes as indicated by the UPDATE operation value (u) and the _hoodie_commit_time = 2023-02-27 03:17:13.915.

We can run the same Hudi Point in Time Query but change the time range to two arbitrary time values, both before the updated record was written (_hoodie_commit_time = 2023-02-26 22:39:07.203). This time, the original record is returned as indicated by the READ operation value (r) and the _hoodie_commit_time of 2023-02-27 03:13:50.642. Again, this is one of the strengths of Apache Hudi, the ability to query data in the present or at any point in the past.

Querying Hudi Tables with Amazon Athena
We can also use Amazon Athena and run a SQL query against the AWS Glue Data Catalog database’s sale table to view the updated database record stored in Amazon S3 using Hudi’s CoW table format. The operation (op) column’s value now indicates an UPDATE (u).

How are Deletes Handled?
In this post’s architecture, deleted database records are signified with an operation ("op") field value of "d" for a DELETE and a __deleted field value of true.
https://itnext.io/media/f7c4ad62dcd45543fdf63834814a5aab
Back to our previous Jupyter notebook example, when rerunning the Spark SQL query, the latest record is returned, reflecting the changes as indicated by the DELETE operation value (d) and the _hoodie_commit_time = 2023-02-27 03:52:13.689.

Alternatively, we could use an additional Spark SQL filter statement to prevent deleted records from being returned (e.g., df.__op != "d").

Since we only did what is referred to as a soft delete in Hudi terminology, we can run a time travel query with an as.of.instance set to some arbitrary time before the deleted record was written (_hoodie_commit_time = 2023-02-27 03:52:13.689). This time, the original record is returned as indicated by the READ operation value (r) and the _hoodie_commit_time = 2023-02-27 03:13:50.642. We could also use a later as.of.instance to return the version of the record reflecting the the UPDATE operation value (u). This also applies to other query types such as point-in-time queries.

Conclusion
In this post, we learned how to build a near real-time transactional data lake on AWS using one possible architecture. The data lake was built using a combination of open source software (OSS) and fully-managed AWS services. Red Hat’s Debezium, Apache Kafka, and Kafka Connect were used for change data capture (CDC). In addition, Apache Spark, Apache Hudi, and Hudi’s DeltaStreamer were used to manage the data lake. To complete our architecture, we used several fully-managed AWS services, including Amazon RDS, Amazon MKS, Amazon EKS, AWS Glue, and Amazon EMR.
This blog represents my own viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners.
Serverless Analytics on AWS: Getting Started with Amazon EMR Serverless and Amazon MSK Serverless
Utilizing the recently released Amazon EMR Serverless and Amazon MSK Serverless for batch and streaming analytics with Apache Spark and Apache Kafka
Introduction
Amazon EMR Serverless
AWS recently announced the general availability (GA) of Amazon EMR Serverless on June 1, 2022. EMR Serverless is a new serverless deployment option in Amazon EMR, in addition to EMR on EC2, EMR on EKS, and EMR on AWS Outposts. EMR Serverless provides a serverless runtime environment that simplifies the operation of analytics applications that use the latest open source frameworks, such as Apache Spark and Apache Hive. According to AWS, with EMR Serverless, you don’t have to configure, optimize, secure, or operate clusters to run applications with these frameworks.
Amazon MSK Serverless
Similarly, on April 28, 2022, AWS announced the general availability of Amazon MSK Serverless. According to AWS, Amazon MSK Serverless is a cluster type for Amazon MSK that makes it easy to run Apache Kafka without managing and scaling cluster capacity. MSK Serverless automatically provisions and scales compute and storage resources, so you can use Apache Kafka on demand and only pay for the data you stream and retain.
Serverless Analytics
In the following post, we will learn how to use these two new, powerful, cost-effective, and easy-to-operate serverless technologies to perform batch and streaming analytics. The PySpark examples used in this post are similar to those featured in two earlier posts, which featured non-serverless alternatives Amazon EMR on EC2 and Amazon MSK: Getting Started with Spark Structured Streaming and Kafka on AWS using Amazon MSK and Amazon EMR and Stream Processing with Apache Spark, Kafka, Avro, and Apicurio Registry on AWS using Amazon MSK and EMR.
Source Code
All the source code demonstrated in this post is open-source and available on GitHub.
git clone --depth 1 -b main \
https://github.com/garystafford/emr-msk-serverless-demo.git
Architecture
The post’s high-level architecture consists of an Amazon EMR Serverless Application, Amazon MSK Serverless Cluster, and Amazon EC2 Kafka client instance. To support these three resources, we will need two Amazon Virtual Private Clouds (VPCs), a minimum of three subnets, an AWS Internet Gateway (IGW) or equivalent, an Amazon S3 Bucket, multiple AWS Identity and Access Management (IAM) Roles and Policies, Security Groups, and Route Tables, and a VPC Gateway Endpoint for S3. All resources are constrained to a single AWS account and a single AWS Region, us-east-1.
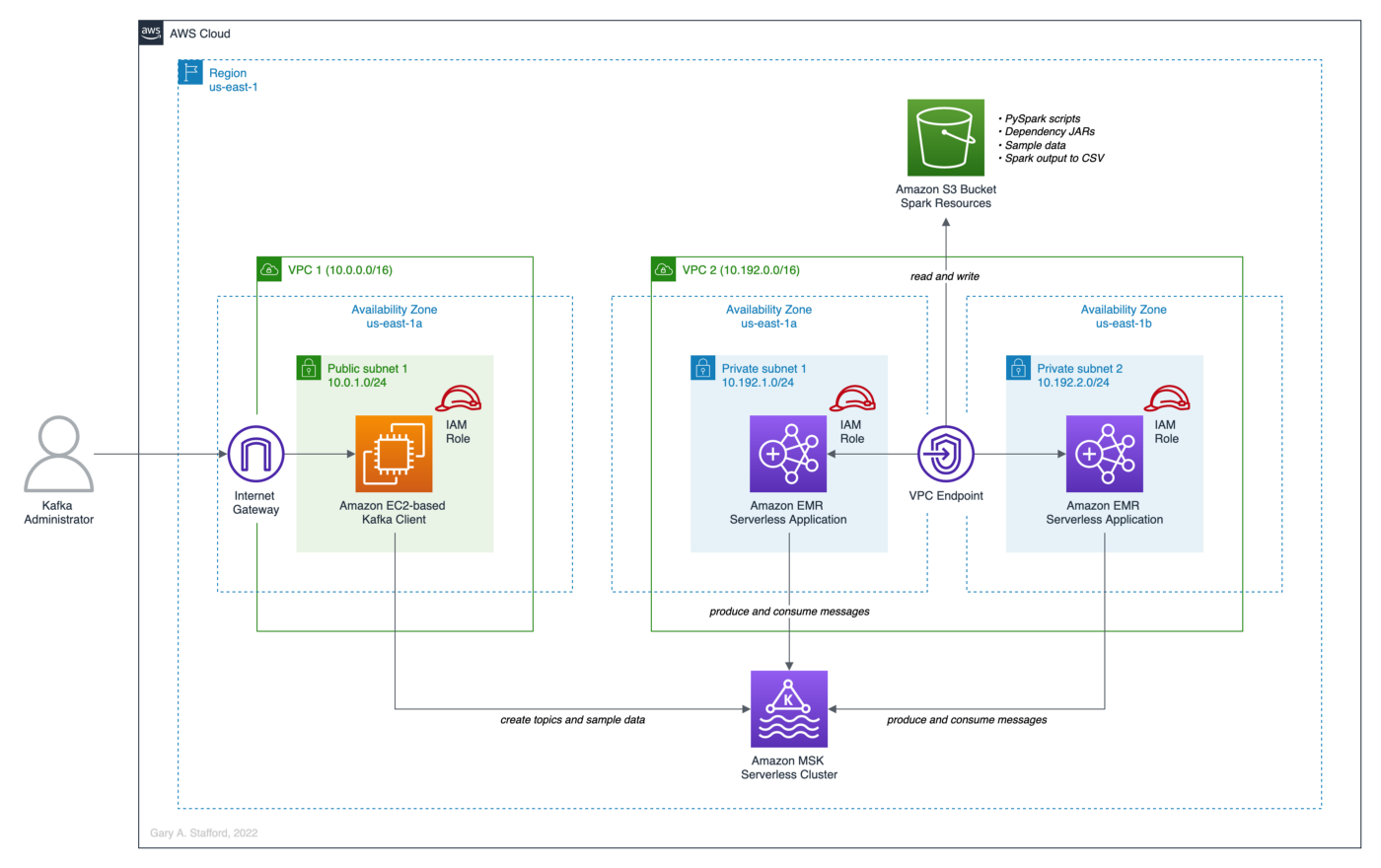
Prerequisites
As a prerequisite for this post, you will need to create the following resources:
- (1) Amazon EMR Serverless Application;
- (1) Amazon MSK Serverless Cluster;
- (1) Amazon S3 Bucket;
- (1) VPC Endpoint for S3;
- (3) Apache Kafka topics;
- PySpark applications, related JAR dependencies, and sample data files uploaded to Amazon S3 Bucket;
Let’s walk through each of these prerequisites.
Amazon EMR Serverless Application
Before continuing, I suggest familiarizing yourself with the AWS documentation for Amazon EMR Serverless, especially, What is Amazon EMR Serverless? Create a new EMR Serverless Application by following the AWS documentation, Getting started with Amazon EMR Serverless. The creation of the EMR Serverless Application includes the following resources:
- Amazon S3 bucket for storage of Spark resources;
- Amazon VPC with at least two private subnets and associated Security Group(s);
- EMR Serverless runtime AWS IAM Role and associated IAM Policy;
- Amazon EMR Serverless Application;
For this post, use the latest version of EMR available in the EMR Studio Serverless Application console, the newly released version 6.7.0, to create a Spark application.

Keep the default pre-initialized capacity, application limits, and application behavior settings.

Since we are connecting to MSK Serverless from EMR Serverless, we need to configure VPC access. Select the new VPC and at least two private subnets in different Availability Zones (AZs).

According to the documentation, the subnets selected for EMR Serverless must be private subnets. The associated route tables for the subnets should not contain direct routes to the Internet.


Amazon MSK Serverless Cluster
Similarly, before continuing, I suggest familiarizing yourself with the AWS documentation for Amazon MSK Serverless, especially MSK Serverless. Create a new MSK Serverless Cluster by following the AWS documentation, Getting started using MSK Serverless clusters. The creation of the MSK Serverless Cluster includes the following resources:
- AWS IAM Role and associated IAM Policy for the Amazon EC2 Kafka client instance;
- VPC with at least one public subnet and associated Security Group(s);
- Amazon EC2 instance used as Apache Kafka client, provisioned in the public subnet of the above VPC;
- Amazon MSK Serverless Cluster;

Associate the new MSK Serverless Cluster with the EMR Serverless Application’s VPC and two private subnets. Also, associate the cluster with the EC2-based Kafka client instance’s VPC and its public subnet.


According to the AWS documentation, Amazon MSK does not support all AZs. For example, I tried to use a subnet in us-east-1e threw an error. If this happens, choose an alternative AZ.


VPC Endpoint for S3
To access the Spark resource in Amazon S3 from EMR Serverless running in the two private subnets, we need a VPC Endpoint for S3. Specifically, a Gateway Endpoint, which sends traffic to Amazon S3 or DynamoDB using private IP addresses. A gateway endpoint for Amazon S3 enables you to use private IP addresses to access Amazon S3 without exposure to the public Internet. EMR Serverless does not require public IP addresses, and you don’t need an internet gateway (IGW), a NAT device, or a virtual private gateway in your VPC to connect to S3.

Create the VPC Endpoint for S3 (Gateway Endpoint) and add the route table for the two EMR Serverless private subnets. You can add additional routes to that route table, such as VPC peering connections to data sources such as Amazon Redshift or Amazon RDS. However, do not add routes that provide direct Internet access.

Kafka Topics and Sample Messages
Once the MSK Serverless Cluster and EC2-based Kafka client instance are provisioned and running, create the three required Kafka topics using the EC2-based Kafka client instance. I recommend using AWS Systems Manager Session Manager to connect to the client instance as the ec2-user user. Session Manager provides secure and auditable node management without the need to open inbound ports, maintain bastion hosts, or manage SSH keys. Alternatively, you can SSH into the client instance.

Before creating the topics, use a utility like telnet to confirm connectivity between the Kafka client and the MSK Serverless Cluster. Verifying connectivity will save you a lot of frustration with potential security and networking issues.
With MSK Serverless Cluster connectivity confirmed, create the three Kafka topics: topicA, topicB, and topicC. I am using the default partitioning and replication settings from the AWS Getting Started Tutorial.
To create some quick sample data, we will copy and paste 250 messages from a file included in the GitHub project, sample_data/sales_messages.txt, into topicA. The messages are simple mock sales transactions.
Use the kafka-console-producer Shell script to publish the messages to the Kafka topic. Use the kafka-console-consumer Shell script to validate the messages made it to the topic by consuming a few messages.
The output should look similar to the following example.

Spark Resources in Amazon S3
To submit and run the five Spark Jobs included in the project, you will need to copy the following resources to your Amazon S3 bucket: (5) Apache Spark jobs, (5) related JAR dependencies, and (2) sample data files.
PySpark Applications
To start, copy the five PySpark applications to a scripts/ subdirectory within your Amazon S3 bucket.

Sample Data
Next, copy the two sample data files to a sample_data/ subdirectory within your Amazon S3 bucket. The large file contains 2,000 messages, while the small file contains 600 messages. These two files can be used interchangeably with the post’s final streaming example.

PySpark Dependencies
Lastly, the PySpark applications have a handful of JAR dependencies that must be available when the job runs, which are not on the EMR Serverless classpath by default. If you are unsure which JARs are already on the EMR Serverless classpath, you can check the Spark UI’s Environment tab’s Classpath Entries section. Accessing the Spark UI is demonstrated in the first PySpark application example, below.

It is critical to choose the correct version of each JAR dependency based on the version of libraries used with the EMR and MSK. Using the wrong version or inconsistent versions, especially Scala, can result in job failures. Specifically, we are targeting Spark 3.2.1 and Scala 2.12 (EMR v6.7.0: Amazon’s Spark 3.2.1, Scala 2.12.15, Amazon Corretto 8 version of OpenJDK), and Apache Kafka 2.8.1 (MSK Serverless: Kafka 2.8.1).
Download the seven JAR files locally, then copy them to a jars/ subdirectory within your Amazon S3 bucket.

PySpark Applications Examples
With the EMR Serverless Application, MSK Serverless Cluster, Kafka topics, and sample data created, and the Spark resources uploaded to Amazon S3, we are ready to explore four different Spark examples.
Example 1: Kafka Batch Aggregation to the Console
The first PySpark application, 01_example_console.py, reads the same 250 sample sales messages from topicA you published earlier, aggregates the messages, and writes the total sales and quantity of orders by country to the console (stdout).
There are no hard-coded values in any of the PySpark application examples. All required environment-specific variables, such as your MSK Serverless bootstrap server (host and port) and Amazon S3 bucket name, will be passed to the running Spark jobs as arguments from the spark-submit command.
To submit your first PySpark job to the EMR Serverless Application, use the emr-serverless API from the AWS CLI. You will need (4) values: 1) your EMR Serverless Application’s application-id, 2) the ARN of your EMR Serverless Application’s execution IAM Role, 3) your MSK Serverless bootstrap server (host and port), and 4) the name of your Amazon S3 bucket containing the Spark resources.
Switching to the EMR Serverless Application console, you should see the new Spark job you just submitted in one of several job states.

You can click on the Spark job to get more details. Note the Script arguments and Spark properties passed in from the spark-submit command.

From the Spark job details tab, access the Spark UI, aka Spark Web UI, from a button in the upper right corner of the screen. If you have experience with Spark, you are most likely familiar with the Spark Web UI to monitor and tune Spark jobs.

From the initial screen, the Spark History Server tab, click on the App ID. You can access an enormous amount of Spark-related information about your job and EMR environment from the Spark Web UI.



The Executors tab will give you access to the Spark job’s output. The output we are most interested in is the driver executor’s stderr and stdout (first row of the second table, shown below).

The stderr contains output related to the running Spark job. Below we see an example of Kafka consumer configuration values output to stderr. Several of these values were passed in from the Spark job, including items such as kafka.bootstrap.servers, security.protocol, sasl.mechanism, and sasl.jaas.config.

The stdout from the driver executor contains the console output as directed from the Spark job. Below we see the successfully aggregated results of the first Spark job, output to stdout.
Example 2: Kafka Batch Aggregation to CSV in S3
Although the console is useful for development and debugging, it is typically not used in Production. Instead, Spark typically sends results to S3 as CSV, JSON, Parquet, or Arvo formatted files, to Kafka, to a database, or to an API endpoint. The second PySpark application, 02_example_csv_s3.py, reads the same 250 sample sales messages from topicA you published earlier, aggregates the messages, and writes the total sales and quantity of orders by country to a CSV file in Amazon S3.
To submit your second PySpark job to the EMR Serverless Application, use the emr-serverless API from the AWS CLI. Similar to the first example, you will need (4) values: 1) your EMR Serverless Application’s application-id, 2) the ARN of your EMR Serverless Application’s execution IAM Role, 3) your MSK Serverless bootstrap server (host and port), and 4) the name of your Amazon S3 bucket containing the Spark resources.
If successful, the Spark job should create a single CSV file in the designated Amazon S3 key (directory path) and an empty _SUCCESS indicator file. The presence of an empty _SUCCESS file signifies that the save() operation completed normally.

Below we see the expected pipe-delimited output from the second Spark job.
Example 3: Kafka Batch Aggregation to Kafka
The third PySpark application, 03_example_kafka.py, reads the same 250 sample sales messages from topicA you published earlier, aggregates the messages, and writes the total sales and quantity of orders by country to a second Kafka topic, topicB. This job now has both read and write options.
To submit your next PySpark job to the EMR Serverless Application, use the emr-serverless API from the AWS CLI. Similar to the first two examples, you will need (4) values: 1) your EMR Serverless Application’s application-id, 2) the ARN of your EMR Serverless Application’s execution IAM Role, 3) your MSK Serverless bootstrap server (host and port), and 4) the name of your Amazon S3 bucket containing the Spark resources.
Once the job completes, you can confirm the results by returning to your EC2-based Kafka client. Use the same kafka-console-consumer command you used previously to show messages from topicB.
If the Spark job and the Kafka client command worked successfully, you should see aggregated messages similar to the example output below. Note we are not using keys with the Kafka messages, only values for these simple examples.

Example 4: Spark Structured Streaming
For our final example, we will switch from batch to streaming — from read to readstream and from write to writestream. Before continuing, I suggest reading the Structured Streaming Programming Guide.
In this example, we will demonstrate how to continuously measure a common business metric — real-time sales volumes. Imagine you are sell products globally and want to understand the relationship between the time of day and buying patterns in different geographic regions in real-time. For any given window of time — this 15-minute period, this hour, this day, or this week— you want to know the current sales volumes by country. You are not reviewing previous sales periods or examing running sales totals, but real-time sales during a sliding time window.
We will use two PySpark jobs running concurrently to simulate this metric. The first application, 04_stream_sales_to_kafka.py, simulates streaming data by continuously writing messages to topicC — 2,000 messages with a 0.5-second delay between messages. In my tests, the job ran for ~28–29 minutes.
Simultaneously, the PySpark application, 05_streaming_kafka.py, continuously consumes the sales transaction messages from the same topic, topicC. Then, Spark aggregates messages over a sliding event-time window and writes the results to the console.
To submit the two PySpark jobs to the EMR Serverless Application, use the emr-serverless API from the AWS CLI. Again, you will need (4) values: 1) your EMR Serverless Application’s application-id, 2) the ARN of your EMR Serverless Application’s execution IAM Role, 3) your MSK Serverless bootstrap server (host and port), and 4) the name of your Amazon S3 bucket containing the Spark resources.
Switching to the EMR Serverless Application console, you should see both Spark jobs you just submitted in one of several job states.

Using the Spark UI again, we can review the output from the second job, 05_streaming_kafka.py.

With Spark Structured Streaming jobs, we have an extra tab in the Spark UI, Structured Streaming. This tab displays all running jobs with their latest [micro]batch number, the aggregate rate of data arriving, and the aggregate rate at which Spark is processing data. Unfortunately, with MSK Serverless, AWS doesn’t appear to allow access to the detailed streaming query statistics via the Run ID, which greatly reduces its value. You receive a 502 error when clicking on the Run ID hyperlink.

The output we are most interested in, again, is contained in the driver executor’s stderr and stdout (first row of the second table, shown below).

Below we see sample output from stderr. The output shows the results of a micro-batch. According to the Apache Spark documentation, internally, by default, Structured Streaming queries are processed using a micro-batch processing engine. The engine processes data streams as a series of small batch jobs, achieving end-to-end latencies as low as 100ms and exactly-once fault-tolerance guarantees.
The corresponding output to the micro-batch output above is shown below. We see the initial micro-batch results, starting with the first micro-batch before any messages are streamed to topicC.
If you are familiar with Spark Structured Streaming, you are likely aware that these Spark jobs run continuously. In other words, the streaming jobs will not stop; they continually await more streaming data.

The first job, 04_stream_sales_to_kafka.py, will run for ~28–29 minutes and stop with a status of Sucess. However, the second job, 05_streaming_kafka.py, the Spark Structured Streaming job, must be manually canceled.

Cleaning Up
You can delete your resources from the AWS Management Console or AWS CLI. However, to delete your Amazon S3 bucket, all objects (including all object versions and delete markers) in the bucket must be deleted before the bucket itself can be deleted.
Conclusion
In this post, we discovered how easy it is to adopt a serverless approach to Analytics on AWS. With EMR Serverless, you don’t have to configure, optimize, secure, or operate clusters to run applications with these frameworks. With MSK Serverless, you can use Apache Kafka on demand and pay for the data you stream and retain. In addition, MSK Serverless automatically provisions and scales compute and storage resources. Given suitable analytics use cases, EMR Serverless with MSK Serverless will likely save you time, effort, and expense.
This blog represents my viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners. All diagrams and illustrations are the property of the author unless otherwise noted.
Developing Spring Boot Applications for Querying Data Lakes on AWS using Amazon Athena
Posted by Gary A. Stafford in Analytics, AWS, Big Data, Cloud, Java Development, Software Development on June 26, 2022
Learn how to develop Cloud-native, RESTful Java services that query data in an AWS-based data lake using Amazon Athena’s API
Introduction
AWS provides a collection of fully-managed services that makes building and managing secure data lakes faster and easier, including AWS Lake Formation, AWS Glue, and Amazon S3. Additional analytics services such as Amazon EMR, AWS Glue Studio, and Amazon Redshift allow Data Scientists and Analysts to run high-performance queries on large volumes of semi-structured and structured data quickly and economically.
What is not always as obvious is how teams develop internal and external customer-facing analytics applications built on top of data lakes. For example, imagine sellers on an eCommerce platform, the scenario used in this post, want to make better marketing decisions regarding their products by analyzing sales trends and buyer preferences. Further, suppose the data required for the analysis must be aggregated from multiple systems and data sources; the ideal use case for a data lake.
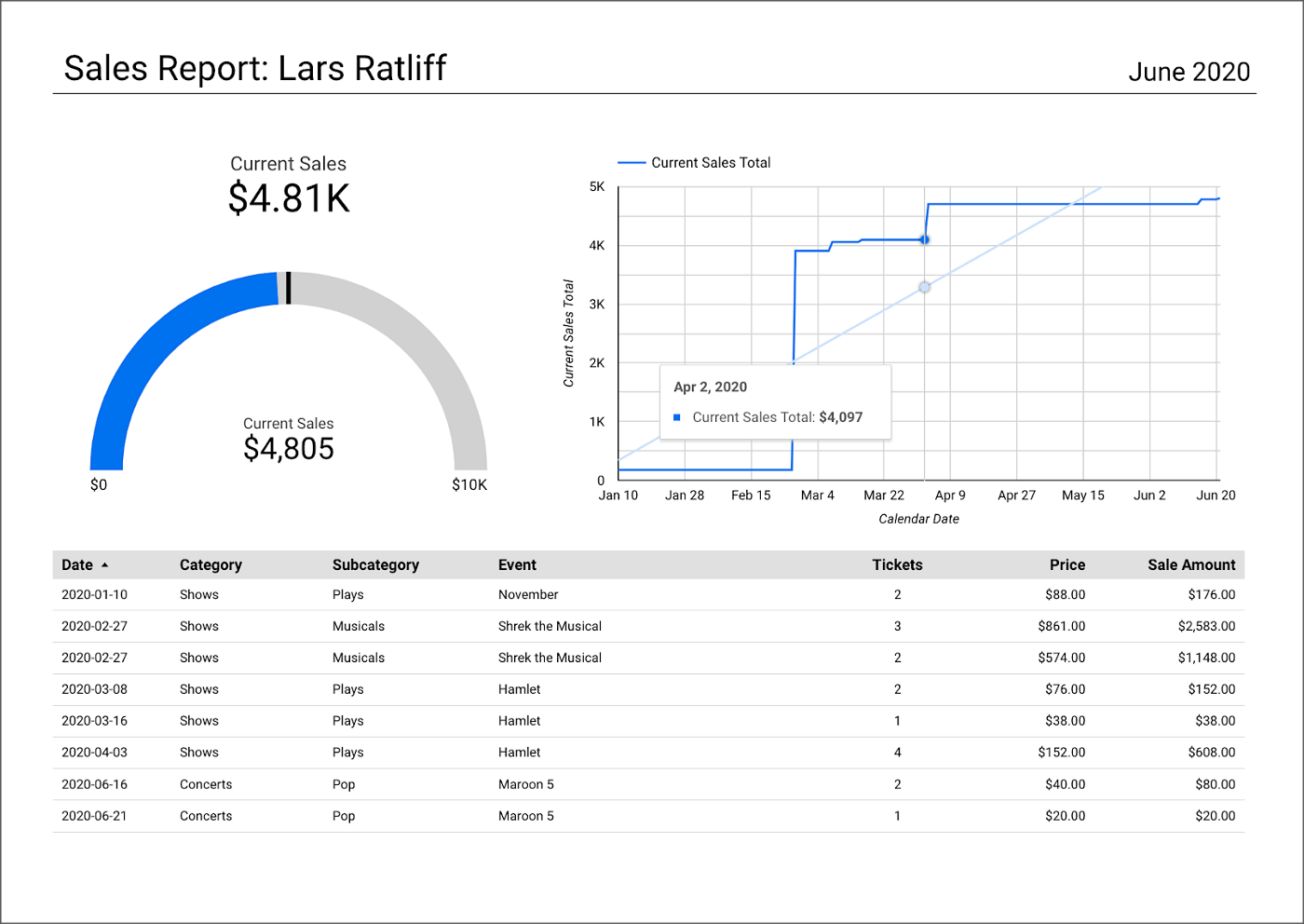
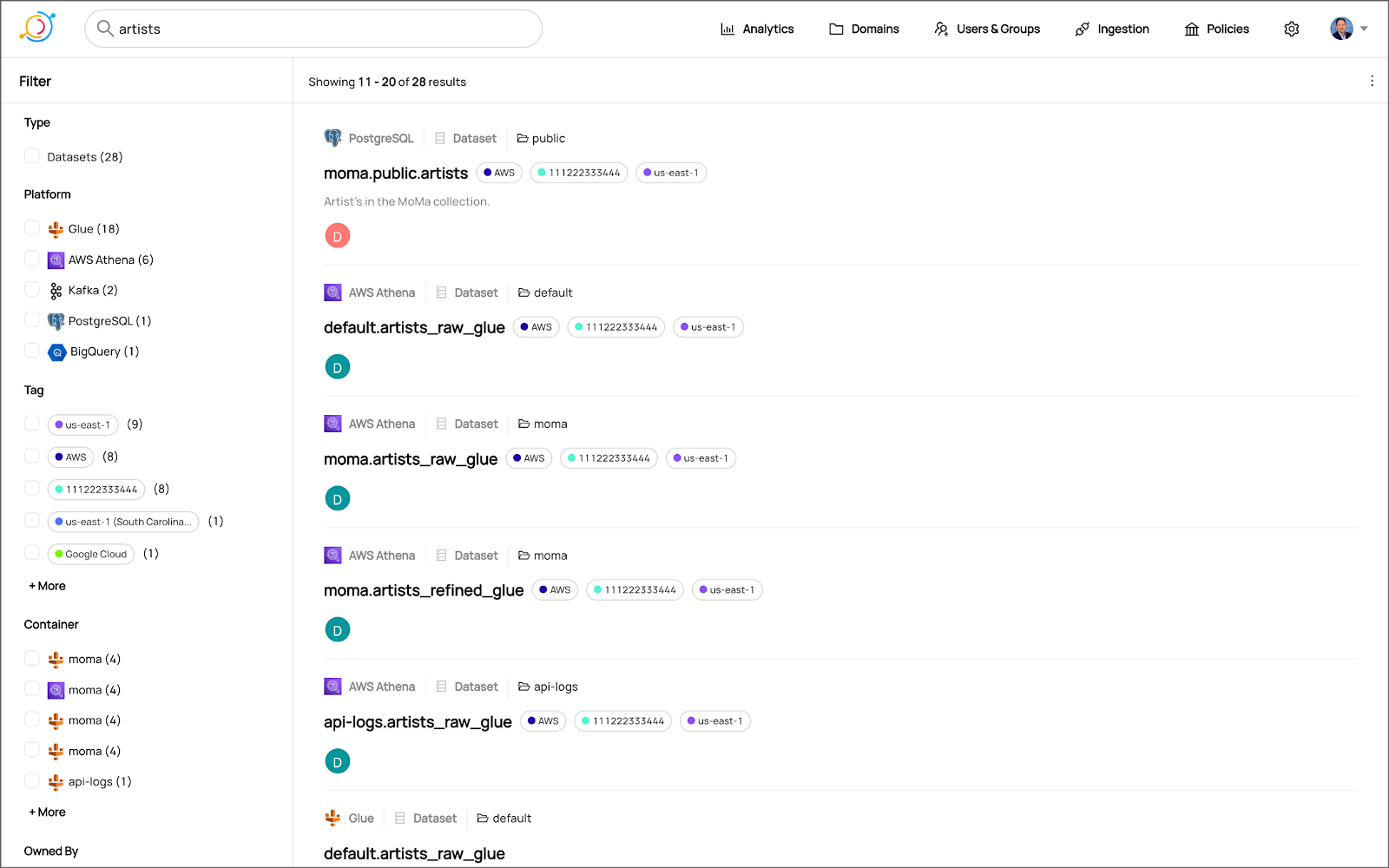
salesbyseller endpointIn this post, we will explore an example Java Spring Boot RESTful Web Service that allows end-users to query data stored in a data lake on AWS. The RESTful Web Service will access data stored as Apache Parquet in Amazon S3 through an AWS Glue Data Catalog using Amazon Athena. The service will use Spring Boot and the AWS SDK for Java to expose a secure, RESTful Application Programming Interface (API).
Amazon Athena is a serverless, interactive query service based on Presto, used to query data and analyze big data in Amazon S3 using standard SQL. Using Athena functionality exposed by the AWS SDK for Java and Athena API, the Spring Boot service will demonstrate how to access tables, views, prepared statements, and saved queries (aka named queries).

TL;DR
Do you want to explore the source code for this post’s Spring Boot service or deploy it to Kubernetes before reading the full article? All the source code, Docker, and Kubernetes resources are open-source and available on GitHub.
git clone --depth 1 -b main \
https://github.com/garystafford/athena-spring-app.git
A Docker image for the Spring Boot service is also available on Docker Hub.

Data Lake Data Source
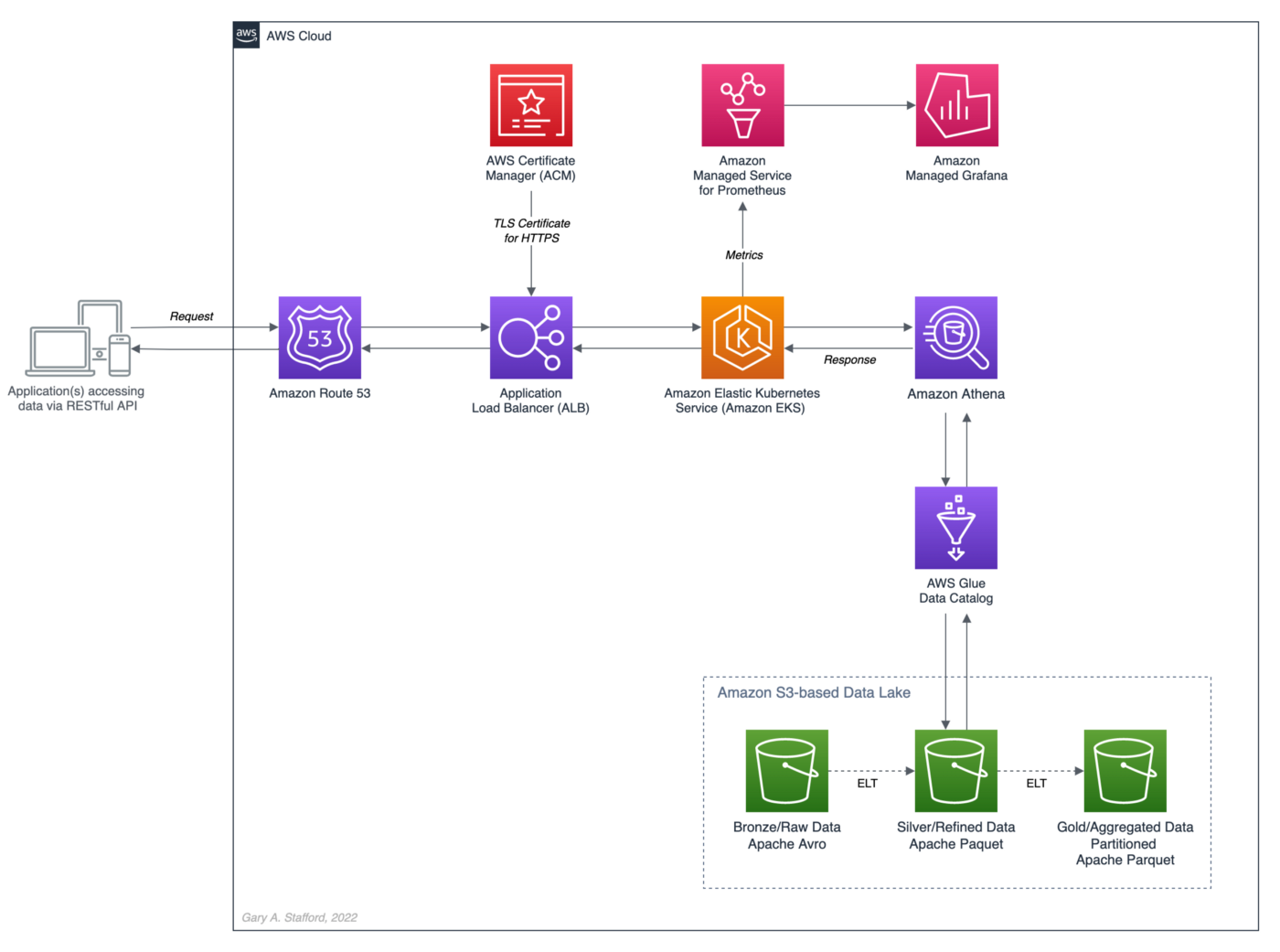
There are endless data sources to build a demonstration data lake on AWS. This post uses the TICKIT sample database provided by AWS and designed for Amazon Redshift, AWS’s cloud data warehousing service. The database consists of seven tables. Two previous posts and associated videos, Building a Data Lake on AWS with Apache Airflow and Building a Data Lake on AWS, detail the setup of the data lake used in this post using AWS Glue and optionally Apache Airflow with Amazon MWAA.
Those two posts use the data lake pattern of segmenting data as bronze (aka raw), silver (aka refined), and gold (aka aggregated), popularized by Databricks. The data lake simulates a typical scenario where data originates from multiple sources, including an e-commerce platform, a CRM system, and a SaaS provider must be aggregated and analyzed.
Spring Projects with IntelliJ IDE
Although not a requirement, I used JetBrains IntelliJ IDEA 2022 (Ultimate Edition) to develop and test the post’s Spring Boot service. Bootstrapping Spring projects with IntelliJ is easy. Developers can quickly create a Spring project using the Spring Initializr plugin bundled with the IntelliJ.

The Spring Initializr plugin’s new project creation wizard is based on start.spring.io. The plugin allows you to quickly select the Spring dependencies you want to incorporate into your project.

Visual Studio Code
There are also several Spring extensions for the popular Visual Studio Code IDE, including Microsoft’s Spring Initializr Java Support extension.

Gradle
This post uses Gradle instead of Maven to develop, test, build, package, and deploy the Spring service. Based on the packages selected in the new project setup shown above, the Spring Initializr plugin’s new project creation wizard creates a build.gradle file. Additional packages, such as Lombak, Micrometer, and Rest Assured, were added separately.
Amazon Corretto
The Spring boot service is developed for and compiled with the most recent version of Amazon Corretto 17. According to AWS, Amazon Corretto is a no-cost, multiplatform, production-ready distribution of the Open Java Development Kit (OpenJDK). Corretto comes with long-term support that includes performance enhancements and security fixes. Corretto is certified as compatible with the Java SE standard and is used internally at Amazon for many production services.
Source Code
Each API endpoint in the Spring Boot RESTful Web Service has a corresponding POJO data model class, service interface and service implementation class, and controller class. In addition, there are also common classes such as configuration, a client factory, and Athena-specific request/response methods. Lastly, there are additional class dependencies for views and prepared statements.

refined_tickit_public_category tableThe project’s source code is arranged in a logical hierarchy by package and class type.
Amazon Athena Access
There are three standard methods for accessing Amazon Athena with the AWS SDK for Java: 1) the AthenaClient service client, 2) the AthenaAsyncClient service client for accessing Athena asynchronously, and 3) using the JDBC driver with the AWS SDK. The AthenaClient and AthenaAsyncClient service clients are both parts of the software.amazon.awssdk.services.athena package. For simplicity, this post’s Spring Boot service uses the AthenaClient service client instead of Java’s asynchronously programming model. AWS supplies basic code samples as part of their documentation as a starting point for writing Athena applications using the SDK. The code samples also use the AthenaClient service client.
POJO-based Data Model Class
For each API endpoint in the Spring Boot RESTful Web Service, there is a corresponding Plain Old Java Object (POJO). According to Wikipedia, a POGO is an ordinary Java object, not bound by any particular restriction. The POJO class is similar to a JPA Entity, representing persistent data stored in a relational database. In this case, the POJO uses Lombok’s @Data annotation. According to the documentation, this annotation generates getters for all fields, a useful toString method, and hashCode and equals implementations that check all non-transient fields. It also generates setters for all non-final fields and a constructor.
Each POJO corresponds directly to a ‘silver’ table in the AWS Glue Data Catalog. For example, the Event POJO corresponds to the refined_tickit_public_event table in the tickit_demo Data Catalog database. The POJO defines the Spring Boot service’s data model for data read from the corresponding AWS Glue Data Catalog table.

The Glue Data Catalog table is the interface between the Athena query and the underlying data stored in Amazon S3 object storage. The Athena query targets the table, which returns the underlying data from S3.

Service Class
Retrieving data from the data lake via AWS Glue, using Athena, is handled by a service class. For each API endpoint in the Spring Boot RESTful Web Service, there is a corresponding Service Interface and implementation class. The service implementation class uses Spring Framework’s @Service annotation. According to the documentation, it indicates that an annotated class is a “Service,” initially defined by Domain-Driven Design (Evans, 2003) as “an operation offered as an interface that stands alone in the model, with no encapsulated state.” Most importantly for the Spring Boot service, this annotation serves as a specialization of @Component, allowing for implementation classes to be autodetected through classpath scanning.
Using Spring’s common constructor-based Dependency Injection (DI) method (aka constructor injection), the service auto-wires an instance of the AthenaClientFactory interface. Note that we are auto-wiring the service interface, not the service implementation, allowing us to wire in a different implementation if desired, such as for testing.
The service calls the AthenaClientFactoryclass’s createClient() method, which returns a connection to Amazon Athena using one of several available authentication methods. The authentication scheme will depend on where the service is deployed and how you want to securely connect to AWS. Some options include environment variables, local AWS profile, EC2 instance profile, or token from the web identity provider.
The service class transforms the payload returned by an instance of GetQueryResultsResponse into an ordered collection (also known as a sequence), List<E>, where E represents a POJO. For example, with the data lake’srefined_tickit_public_event table, the service returns a List<Event>. This pattern repeats itself for tables, views, prepared statements, and named queries. Column data types can be transformed and formatted on the fly, new columns added, and existing columns skipped.
For each endpoint defined in the Controller class, for example, get(), findAll(), and FindById(), there is a corresponding method in the Service class. Below, we see an example of the findAll() method in the SalesByCategoryServiceImp service class. This method corresponds to the identically named method in the SalesByCategoryController controller class. Each of these service methods follows a similar pattern of constructing a dynamic Athena SQL query based on input parameters, which is passed to Athena through the AthenaClient service client using an instance of GetQueryResultsRequest.
Controller Class
Lastly, there is a corresponding Controller class for each API endpoint in the Spring Boot RESTful Web Service. The controller class uses Spring Framework’s @RestController annotation. According to the documentation, this annotation is a convenience annotation that is itself annotated with @Controller and @ResponseBody. Types that carry this annotation are treated as controllers where @RequestMapping methods assume @ResponseBody semantics by default.
The controller class takes a dependency on the corresponding service class application component using constructor-based Dependency Injection (DI). Like the service example above, we are auto-wiring the service interface, not the service implementation.
The controller is responsible for serializing the ordered collection of POJOs into JSON and returning that JSON payload in the body of the HTTP response to the initial HTTP request.
Querying Views
In addition to querying AWS Glue Data Catalog tables (aka Athena tables), we also query views. According to the documentation, a view in Amazon Athena is a logical table, not a physical table. Therefore, the query that defines a view runs each time the view is referenced in a query.
For convenience, each time the Spring Boot service starts, the main AthenaApplication class calls the View.java class’s CreateView() method to check for the existence of the view, view_tickit_sales_by_day_and_category. If the view does not exist, it is created and becomes accessible to all application end-users. The view is queried through the service’s /salesbycategory endpoint.

This confirm-or-create pattern is repeated for the prepared statement in the main AthenaApplication class (detailed in the next section).
Below, we see the View class called by the service at startup.
Aside from the fact the /salesbycategory endpoint queries a view, everything else is identical to querying a table. This endpoint uses the same model-service-controller pattern.
Executing Prepared Statements
According to the documentation, you can use the Athena parameterized query feature to prepare statements for repeated execution of the same query with different query parameters. The prepared statement used by the service, tickit_sales_by_seller, accepts a single parameter, the ID of the seller (sellerid). The prepared statement is executed using the /salesbyseller endpoint. This scenario simulates an end-user of the analytics application who wants to retrieve enriched sales information about their sales.
The pattern of querying data is similar to tables and views, except instead of using the common SELECT...FROM...WHERE SQL query pattern, we use the EXECUTE...USING pattern.
For example, to execute the prepared statement for a seller with an ID of 3, we would use EXECUTE tickit_sales_by_seller USING 3;. We pass the seller’s ID of 3 as a path parameter similar to other endpoints exposed by the service: /v1/salesbyseller/3.

Again, aside from the fact the /salesbyseller endpoint executes a prepared statement and passes a parameter; everything else is identical to querying a table or a view, using the same model-service-controller pattern.
Working with Named Queries
In addition to tables, views, and prepared statements, Athena has the concept of saved queries, referred to as named queries in the Athena API and when using AWS CloudFormation. You can use the Athena console or API to save, edit, run, rename, and delete queries. The queries are persisted using a NamedQueryId, a unique identifier (UUID) of the query. You must reference the NamedQueryId when working with existing named queries.

There are multiple ways to use and reuse existing named queries programmatically. For this demonstration, I created the named query, buyer_likes_by_category, in advance and then stored the resulting NamedQueryId as an application property, injected at runtime or kubernetes deployment time through a local environment variable.
Alternately, you might iterate through a list of named queries to find one that matches the name at startup. However, this method would undoubtedly impact service performance, startup time, and cost. Lastly, you could use a method like NamedQuery() included in the unused NamedQuery class at startup, similar to the view and prepared statement. That named query’s unique NamedQueryId would be persisted as a system property, referencable by the service class. The downside is that you would create a duplicate of the named query each time you start the service. Therefore, this method is also not recommended.
Configuration
Two components responsible for persisting configuration for the Spring Boot service are the application.yml properties file and ConfigProperties class. The class uses Spring Framework’s @ConfigurationProperties annotation. According to the documentation, this annotation is used for externalized configuration. Add this to a class definition or a @Bean method in a @Configuration class if you want to bind and validate some external Properties (e.g., from a .properties or .yml file). Binding is performed by calling setters on the annotated class or, if @ConstructorBinding in use, by binding to the constructor parameters.
The @ConfigurationProperties annotation includes the prefix of athena. This value corresponds to the athena prefix in the the application.yml properties file. The fields in the ConfigProperties class are bound to the properties in the the application.yml. For example, the property, namedQueryId, is bound to the property, athena.named.query.id. Further, that property is bound to an external environment variable, NAMED_QUERY_ID. These values could be supplied from an external configuration system, a Kubernetes secret, or external secrets management system.
AWS IAM: Authentication and Authorization
For the Spring Boot service to interact with Amazon Athena, AWS Glue, and Amazon S3, you need to establish an AWS IAM Role, which the service assumes once authenticated. The Role must be associated with an attached IAM Policy containing the requisite Athena, Glue, and S3 permissions. For development, the service uses a policy similar to the one shown below. Please note this policy is broader than recommended for Production; it does not represent the security best practice of least privilege. In particular, the use of the overly-broad * for Resources should be strictly avoided when creating policies.
In addition to the authorization granted by the IAM Policy, AWS Lake Formation can be used with Amazon S3, AWS Glue, and Amazon Athena to grant fine-grained database-, table-, column-, and row-level access to datasets.
Swagger UI and the OpenAPI Specification
The easiest way to view and experiment with all the endpoints available through the controller classes is using the Swagger UI, included in the example Spring Boot service, by way of the springdoc-openapi Java library. The Swagger UI is accessed at /v1/swagger-ui/index.html.

The OpenAPI Specification (formerly Swagger Specification) is an API description format for REST APIs. The /v1/v3/api-docs endpoint allows you to generate an OpenAPI v3 specification file. The OpenAPI file describes the entire API.

The OpenAPI v3 specification can be saved as a file and imported into applications like Postman, the API platform for building and using APIs.

/users API endpoint using Postman
Integration Tests
Included in the Spring Boot service’s source code is a limited number of example integration tests, not to be confused with unit tests. Each test class uses Spring Framework’s @SpringBootTest annotation. According to the documentation, this annotation can be specified on a test class that runs Spring Boot-based tests. It provides several features over and above the regular Spring TestContext Framework.
The integration tests use Rest Assured’s given-when-then pattern of testing, made popular as part of Behavior-Driven Development (BDD). In addition, each test uses the JUnit’s @Test annotation. According to the documentation, this annotation signals that the annotated method is a test method. Therefore, methods using this annotation must not be private or static and must not return a value.
Run the integration tests using Gradle from the project’s root: ./gradlew clean build test. A detailed ‘Test Summary’ is produced in the project’s build directory as HTML for easy review.


Load Testing the Service
In Production, the Spring Boot service will need to handle multiple concurrent users executing queries against Amazon Athena.

We could use various load testing tools to evaluate the service’s ability to handle multiple concurrent users. One of the simplest is my favorite go-based utility, hey, which sends load to a URL using a provided number of requests in the provided concurrency level and prints stats. It also supports HTTP2 endpoints. So, for example, we could execute 500 HTTP requests with a concurrency level of 25 against the Spring Boot service’s /users endpoint using hey. The post’s integration tests were run against three Kubernetes replica pods of the service deployed to Amazon EKS.
hey -n 500 -c 25 -T "application/json;charset=UTF-8" \
-h2 https://athena.example-api.com/v1/users

From Athena’s Recent Queries console, we see many simultaneous queries being queued and executed by a hey through the Spring Boot service’s endpoint.

Metrics
The Spring Boot service implements the micrometer-registry-prometheus extension. The Micrometer metrics library exposes runtime and application metrics. Micrometer defines a core library, providing a registration mechanism for metrics and core metric types. These metrics are exposed by the service’s /v1/actuator/prometheus endpoint.

Using the Micrometer extension, metrics exposed by the /v1/actuator/prometheus endpoint can be scraped and visualized by tools such as Prometheus. Conveniently, AWS offers the fully-managed Amazon Managed Service for Prometheus (AMP), which easily integrates with Amazon EKS.

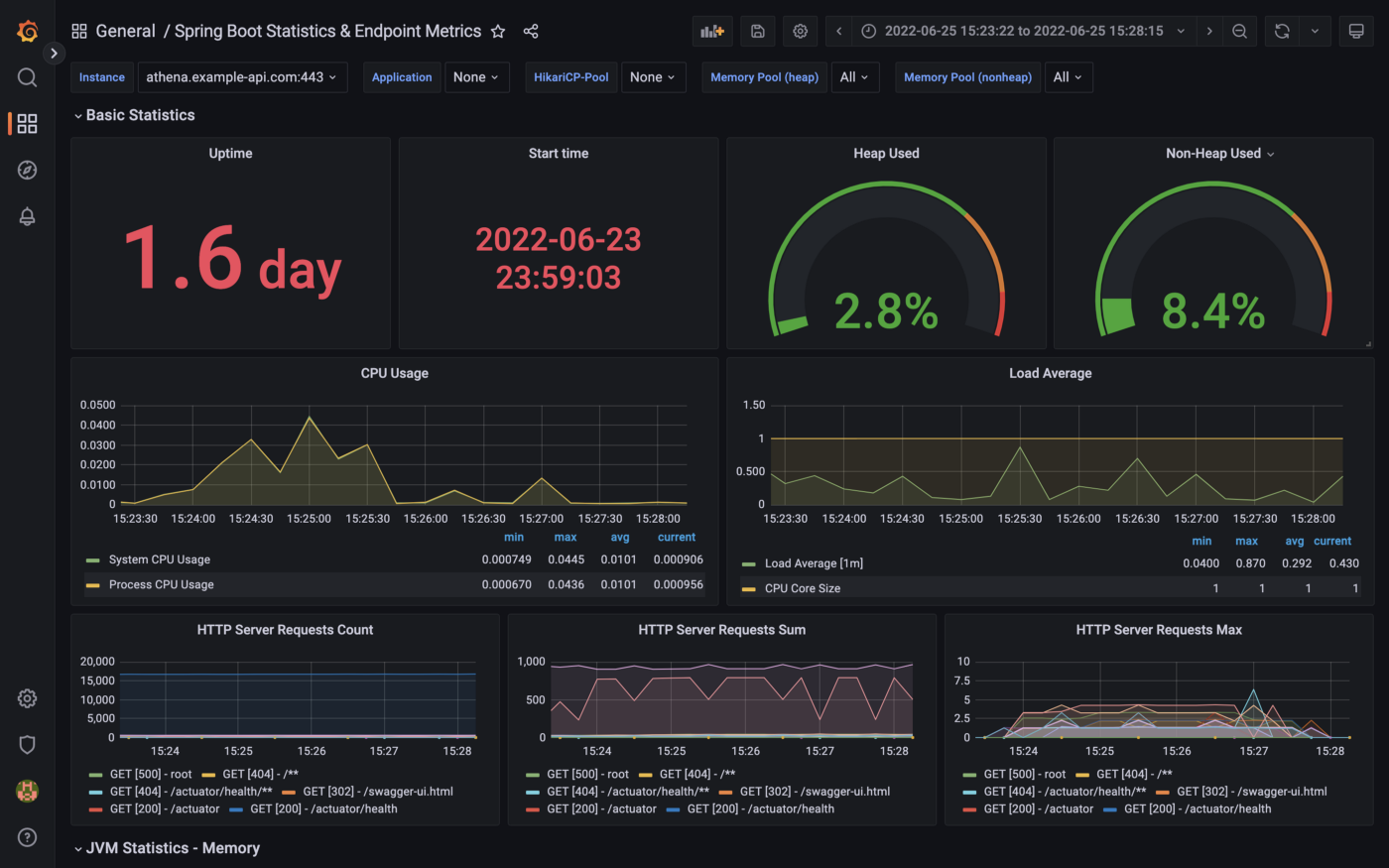
Using Prometheus as a datasource, we can build dashboards in Grafana to observe the service’s metrics. Like AMP, AWS also offers the fully-managed Amazon Managed Grafana (AMG).

Conclusion
This post taught us how to create a Spring Boot RESTful Web Service, allowing end-user applications to securely query data stored in a data lake on AWS. The service used AWS SDK for Java to access data stored in Amazon S3 through an AWS Glue Data Catalog using Amazon Athena.
This blog represents my own viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners. All diagrams and illustrations are property of the author unless otherwise noted.
End-to-End Data Discovery, Observability, and Governance on AWS with LinkedIn’s Open-source DataHub
Posted by Gary A. Stafford in Analytics, AWS, Azure, Bash Scripting, Build Automation, Cloud, DevOps, GCP, Kubernetes, Python, Software Development, SQL, Technology Consulting on March 26, 2022
Use DataHub’s data catalog capabilities to collect, organize, enrich, and search for metadata across multiple platforms
Introduction
According to Shirshanka Das, Founder of LinkedIn DataHub, Apache Gobblin, and Acryl Data, one of the simplest definitions for a data catalog can be found on the Oracle website: “Simply put, a data catalog is an organized inventory of data assets in the organization. It uses metadata to help organizations manage their data. It also helps data professionals collect, organize, access, and enrich metadata to support data discovery and governance.”
Another succinct description of a data catalog’s purpose comes from Alation: “a collection of metadata, combined with data management and search tools, that helps analysts and other data users to find the data that they need, serves as an inventory of available data, and provides information to evaluate the fitness of data for intended uses.”
Working with many organizations in the area of Analytics, one of the more common requests I receive regards choosing and implementing a data catalog. Organizations have datasources hosted in corporate data centers, on AWS, by SaaS providers, and with other Cloud Service Providers. Several of these organizations have recently gravitated to DataHub, the open-source metadata platform for the modern data stack, originally developed by LinkedIn.

In this post, we will explore the capabilities of DataHub to build a centralized data catalog on AWS for datasources hosted in multiple AWS accounts, SaaS providers, cloud service providers, and corporate data centers. I will demonstrate how to build a DataHub data catalog using out-of-the-box data source plugins for automated metadata ingestion.
Data Catalog Competitors
Data catalogs are not new; technologies such as data dictionaries have been around as far back as the 1980’s. Gartner publishes their Metadata Management (EMM) Solutions Reviews and Ratings and Metadata Management Magic Quadrant. These reports contain a comprehensive list of traditional commercial enterprise players, modern cloud-native SaaS vendors, and Cloud Service Provider (CSP) offerings. DBMS Tools also hosts a comprehensive list of 30 data catalogs. A sampling of current data catalogs includes:
Open Source Software
Commercial
- Acryl Data (based on LinkedIn’s DataHub)
- Atlan
- Stemma (based on Lyft’s Amundsen)
- Talend
- Alation
- Collibra
- data.world
Cloud Service Providers
Data Catalog Features
DataHub describes itself as “a modern data catalog built to enable end-to-end data discovery, data observability, and data governance.” Sorting through vendor’s marketing jargon and hype, standard features of leading data catalogs include:
- Metadata ingestion
- Data discovery
- Data governance
- Data observability
- Data lineage
- Data dictionary
- Data classification
- Usage/popularity statistics
- Sensitive data handling
- Data fitness (aka data quality or data profiling)
- Manage both technical and business metadata
- Business glossary
- Tagging
- Natively supported datasource integrations
- Advanced metadata search
- Fine-grain authentication and authorization
- UI- and API-based interaction
Datasources
When considering a data catalog solution, in my experience, the most common datasources that customers want to discover, inventory, and search include:
- Relational databases and other OLTP datasources such as PostgreSQL, MySQL, Microsoft SQL Server, and Oracle
- Cloud Data Warehouses and other OLAP datasources such as Amazon Redshift, Snowflake, and Google BigQuery
- NoSQL datasources such as MongoDB, MongoDB Atlas, and Azure Cosmos DB
- Persistent event-streaming platforms such as Apache Kafka (Amazon MSK and Confluent)
- Distributed storage datasets (e.g., Data Lakes) such as Amazon S3, Apache Hive, and AWS Glue Data Catalogs
- Business Intelligence (BI), dashboards, and data visualization sources such as Looker, Tableau, and Microsoft Power BI
- ETL sources, such as Apache Spark, Apache Airflow, Apache NiFi, and dbt
DataHub on AWS
DataHub’s convenient AWS setup guide covers options to deploy DataHub to AWS. For this post, I have hosted DataHub on Kubernetes, using Amazon Elastic Kubernetes Service (Amazon EKS). Alternately, you could choose Google Kubernetes Engine (GKE) on Google Cloud or Azure Kubernetes Service (AKS) on Microsoft Azure.
Conveniently, DataHub offers a Helm chart, making deployment to Kubernetes straightforward. Furthermore, Helm charts are easily integrated with popular CI/CD tools. For this post, I’ve used ArgoCD, the declarative GitOps continuous delivery tool for Kubernetes, to deploy the DataHub Helm charts to Amazon EKS.

According to the documentation, DataHub consists of four main components: GMS, MAE Consumer (optional), MCE Consumer (optional), and Frontend. Kubernetes deployment for each of the components is defined as sub-charts under the main DataHub Helm chart.
External Storage Layer Dependencies
Four external storage layer dependencies power the main DataHub components: Kafka, Local DB (MySQL, Postgres, or MariaDB), Search Index (Elasticsearch), and Graph Index (Neo4j or Elasticsearch). DataHub has provided a separate DataHub Prerequisites Helm chart for the dependencies. The dependencies must be deployed before deploying DataHub.
Alternately, you can substitute AWS managed services for the external storage layer dependencies, which is also detailed in the Deploying to AWS documentation. AWS managed service dependency substitutions include Amazon RDS for MySQL, Amazon OpenSearch (fka Amazon Elasticsearch), and Amazon Managed Streaming for Apache Kafka (Amazon MSK). According to DataHub, support for using AWS Neptune as the Graph Index is coming soon.
DataHub CLI and Plug-ins
DataHub comes with the datahub CLI, allowing you to perform many common operations on the command line. You can install and use the DataHub CLI within your development environment or integrate it with your CI/CD tooling.

DataHub uses a plugin architecture. Plugins allow you to install only the datasource dependencies you need. For example, if you want to ingest metadata from Amazon Athena, just install the Athena plugin: pip install 'acryl-datahub[athena]'. DataHub Source, Sink, and Transformer plugins can be displayed using the datahub check plugins CLI command.


Secure Metadata Ingestion
Often, datasources are not externally accessible for security reasons. Further, many datasources may not be accessible to individual users, especially in higher environments like UAT, Staging, and Production. They are only accessible to applications or CI/CD tooling. To overcome these limitations when extracting metadata with DataHub, I prefer to perform my DataHub-related development and testing locally but execute all DataHub ingestion securely on AWS.
In my local development environment, I use JetBrains PyCharm to author the Python and YAML-based DataHub configuration files and ingestion pipeline recipes, then commit those files to git and push them to a private GitHub repository. Finally, I use GitHub Actions to test DataHub files.
To run DataHub ingestion jobs and push the results to DataHub running in Kubernetes on Amazon EKS, I have built a custom Python-based Docker container. The container runs the DataHub CLI, required DataHub plugins, and any additional Python dependencies. The container’s pod has the appropriate AWS IAM permissions, using IAM Roles for Service Accounts (IRSA), to securely access datasources to ingest and the DataHub application.
Schedule and Monitor Pipelines
Scheduling and managing multiple metadata ingestion jobs on AWS is best handled with Apache Airflow with Amazon Managed Workflows for Apache Airflow (Amazon MWAA). Ingestion jobs run as Airflow DAG tasks, which call the EKS-based DataHub CLI container. With MWAA, datasource connections, credentials, and other sensitive configurations can be kept secure and not be exposed externally or in plain text.
When running the ingestion pipelines on AWS with DataHub, all communications between AWS-based datasources, ingestion jobs running in Airflow, and DataHub, should use secure private IP addressing and DNS resolution instead of transferring metadata over the Internet. Make sure to create all the necessary VPC peering connections, network route table configurations, and VPC endpoints to connect all relevant services.
SaaS services such as Snowflake or MongoDB Atlas, services provided by other Cloud Service Providers such as Google Cloud and Microsoft Azure, and datasources in corporate datasources require alternate networking and security strategies to access metadata securely.

Markup or Code?
According to the documentation, a DataHub recipe is a configuration file that tells ingestion scripts where to pull data from (source) and where to put it (sink). Recipes normally contain a source, sink, and transformers configuration section. Mark-up language-based job automation written in YAML, JSON, or Domain Specific Languages (DSLs) is often an alternative to writing code. DataHub recipes can be written in YAML. The example recipe shown below is used to ingest metadata from an Amazon RDS for PostgreSQL database, running on AWS.
YAML-based recipes can also use automatic environment variable expansion for convenience, automation, and security. It is considered best practice to secure sensitive configuration values, such as database credentials, in a secure location and reference them as environment variables. For example, note the server: ${DATAHUB_REST_ENDPOINT} entry in the sink section below. The DATAHUB_REST_ENDPOINT environment variable is set ahead of time and re-used for all ingestion jobs. Sensitive database connection information has also been variablized and stored separately.
Using Python
You can configure and run a pipeline entirely from within a custom Python script using DataHub’s Python API as an alternative to YAML. Below, we see two nearly identical ingestion recipes to the YAML above, written in Python. Writing ingestion pipeline logic programmatically gives you increased flexibility for automation, error checking, unit-testing, and notification. Below is a basic pipeline written in Python. The code is functional, but not very Pythonic, secure, scalable, or Production ready.
The second version of the same pipeline is more Production ready. The code is more Pythonic in nature and makes use of error checking, logging, and the AWS Systems Manager (SSM) Parameter Store. Like recipes written in YAML, environment variables can be used for convenience and security. In this example, commonly reused and sensitive connection configuration items have been extracted and placed in the SSM Parameter Store. Additional configuration is pulled from the environment, such as AWS Account ID and AWS Region. The script loads these values at runtime.
Sinking to DataHub
When syncing metadata to DataHub, you have two choices, the GMS REST API or Kafka. According to DataHub, the advantage of the REST-based interface is that any errors can immediately be reported. On the other hand, the advantage of the Kafka-based interface is that it is asynchronous and can handle higher throughput. For this post, I am DataHub’s REST API.


Column-level Metadata
In addition to column names and data types, it is possible to extract column descriptions and key types from certain datasources. Column descriptions, tags, and glossary terms can also be input through the DataHub UI. Below, we see an example of an Amazon Redshift fact table, whose table and column descriptions were ingested as part of the metadata.

Business Glossary
DataHub can assign business glossary terms to entities. The DataHub Business Glossary plugin pulls business glossary metadata from a YAML-based configuration file.
Business glossary terms can be reviewed in the Glossary Terms tab of the DataHub’s UI. Below, we see the three terms associated with the Classification glossary node: Confidential, HighlyConfidential, and Sensitive.

We can search for entities inventoried in DataHub using their assigned business glossary terms.

Finally, we see an example of an AWS Athena data catalog table with business glossary terms applied to columns within the table’s schema.

SQL-based Profiler
DataHub also can extract statistics about entities in DataHub using the SQL-based Profiler. According to the DataHub documentation, the Profiler can extract the following:
- Row and column counts for each table
- Column null counts and proportions
- Column distinct counts and proportions
- Column min, max, mean, median, standard deviation, quantile values
- Column histograms or frequencies of unique values
In addition, we can also track the historical stats for each profiled entity each time metadata is ingested.


Data Lineage
DataHub’s data lineage features allow us to view upstream and downstream relationships between different types of entities. DataHub can trace lineage across multiple platforms, datasets, pipelines, charts, and dashboards.
Below, we see a simple example of dataset entity-to-entity lineage in Amazon Redshift and then Apache Spark on Amazon EMR. The fact table has a downstream relationship to four database views. The views are based on SQL queries that include the upstream table as a datasource.


DataHub Analytics
DataHub provides basic metadata quality and usage analytics in the DataHub UI: user activity, counts of datasource types, business glossary terms, environments, and actions.


Conclusion
In this post, we explored the features of a data catalog and learned about some of the leading commercial and open-source data catalogs. Next, we learned how DataHub could collect, organize, enrich, and search metadata across multiple datasources. Lastly, we discovered how easy it is to catalog metadata from datasources spread across multiple CSP, SaaS providers, and corporate data centers, and centralize those results in DataHub.
In addition to the basic features reviewed in this post, DataHub offers a growing number of additional capabilities, including GraphQL and Timeline APIs, robust authentication and authorization, application monitoring observability, and Great Expectations integration. All these qualities make DataHub an excellent choice for a data catalog.
This blog represents my own viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners.
Data Preparation on AWS: Comparing Available ELT Options to Cleanse and Normalize Data
Posted by Gary A. Stafford in Analytics, AWS, Build Automation, Cloud, Python, SQL, Technology Consulting on March 1, 2022
Comparing the features and performance of different AWS analytics services for Extract, Load, Transform (ELT)
Introduction
According to Wikipedia, “Extract, load, transform (ELT) is an alternative to extract, transform, load (ETL) used with data lake implementations. In contrast to ETL, in ELT models the data is not transformed on entry to the data lake but stored in its original raw format. This enables faster loading times. However, ELT requires sufficient processing power within the data processing engine to carry out the transformation on demand, to return the results in a timely manner.”
As capital investments and customer demand continue to drive the growth of the cloud-based analytics market, the choice of tools seems endless, and that can be a problem. Customers face a constant barrage of commercial and open-source tools for their batch, streaming, and interactive exploratory data analytics needs. The major Cloud Service Providers (CSPs) have even grown to a point where they now offer multiple services to accomplish similar analytics tasks.
This post will examine the choice of analytics services available on AWS capable of performing ELT. Specifically, this post will compare the features and performance of AWS Glue Studio, Amazon Glue DataBrew, Amazon Athena, and Amazon EMR using multiple ELT use cases and service configurations.
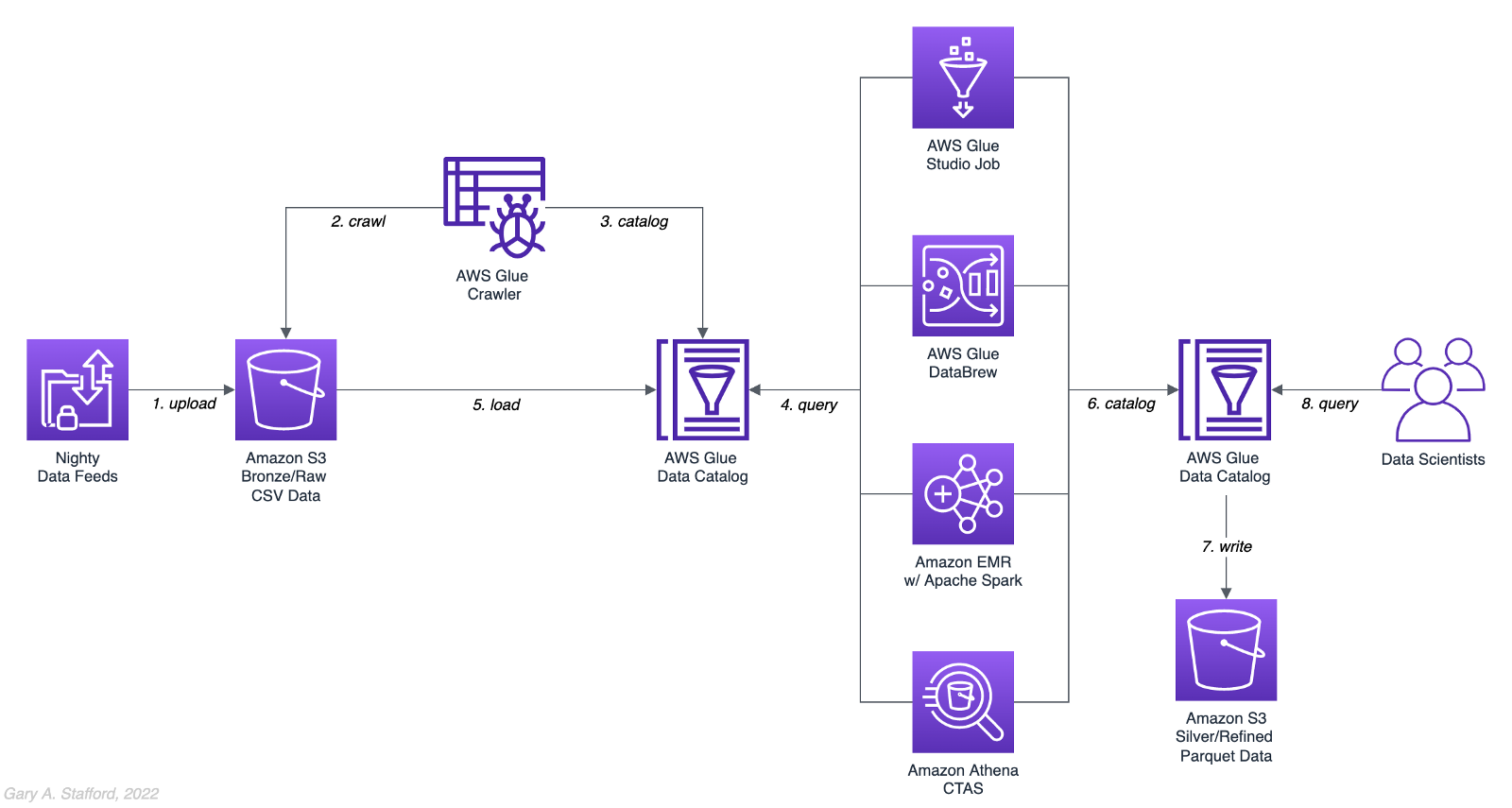
Analytics Use Case
We will address a simple yet common analytics challenge for this comparison — preparing a nightly data feed for analysis the next day. Each night a batch of approximately 1.2 GB of raw CSV-format healthcare data will be exported from a Patient Administration System (PAS) and uploaded to Amazon S3. The data must be cleansed, deduplicated, refined, normalized, and made available to the Data Science team the following morning. The team of Data Scientists will perform complex data analytics on the data and build machine learning models designed for early disease detection and prevention.
Sample Dataset
The dataset used for this comparison is generated by Synthea, an open-source patient population simulation. The high-quality, synthetic, realistic patient data and associated health records cover every aspect of healthcare. The dataset contains the patient-related healthcare history for allergies, care plans, conditions, devices, encounters, imaging studies, immunizations, medications, observations, organizations, patients, payers, procedures, providers, and supplies.
The Synthea dataset was first introduced in my March 2021 post examining the handling of sensitive PII data using Amazon Macie: Data Lakes: Discovery, Security, and Privacy of Sensitive Data.
The Synthea synthetic patient data is available in different record volumes and various data formats, including HL7 FHIR, C-CDA, and CSV. We will use CSV-format data files for this post. Since this post seeks to measure the performance of different AWS ELT-capable services, we will use a larger version of the Synthea dataset containing hundreds of thousands to millions of records.
AWS Glue Data Catalog
The dataset comprises nine uncompressed CSV files uploaded to Amazon S3 and cataloged to an AWS Glue Data Catalog, a persistent metadata store, using an AWS Glue Crawler.

Test Cases
We will use three data preparation test cases based on the Synthea dataset to examine the different AWS ELT-capable services.

Test Case 1: Encounters for Symptom
An encounter is a health care contact between the patient and the provider responsible for diagnosing and treating the patient. In our first test case, we will process 1.26M encounters records for an ongoing study of patient symptoms by our Data Science team.
Data preparation includes the following steps:
- Load 1.26M encounter records using the existing AWS Glue Data Catalog table.
- Remove any duplicate records.
- Select only the records where the
descriptioncolumn contains “Encounter for symptom.” - Remove any rows with an empty
reasoncodescolumn. - Extract a new
year,month, anddaycolumn from thedatecolumn. - Remove the
datecolumn. - Write resulting dataset back to Amazon S3 as Snappy-compressed Apache Parquet files, partitioned by
year,month, andday. - Given the small resultset, bucket the data such that only one file is written per
daypartition to minimize the impact of too many small files on future query performance. - Catalog resulting dataset to a new table in the existing AWS Glue Data Catalog, including partitions.
Test Case 2: Observations
Clinical observations ensure that treatment plans are up-to-date and correctly administered and allow healthcare staff to carry out timely and regular bedside assessments. We will process 5.38M encounters records for our Data Science team in our second test case.
Data preparation includes the following steps:
- Load 5.38M observation records using the existing AWS Glue Data Catalog table.
- Remove any duplicate records.
- Extract a new
year,month, anddaycolumn from the date column. - Remove the
datecolumn. - Write resulting dataset back to Amazon S3 as Snappy-compressed Apache Parquet files, partitioned by
year,month, andday. - Given the small resultset, bucket the data such that only one file is written per
daypartition to minimize the impact of too many small files on future query performance. - Catalog resulting dataset to a new table in the existing AWS Glue Data Catalog, including partitions.
Test Case 3: Sinusitis Study
A medical condition is a broad term that includes all diseases, lesions, and disorders. In our second test case, we will join the conditions records with the patient records and filter for any condition containing the term ‘sinusitis’ in preparation for our Data Science team.
Data preparation includes the following steps:
- Load 483k condition records using the existing AWS Glue Data Catalog table.
- Inner join the condition records with the 132k patient records based on patient ID.
- Remove any duplicate records.
- Drop approximately 15 unneeded columns.
- Select only the records where the
descriptioncolumn contains the term “sinusitis.” - Remove any rows with empty
ethnicity,race,gender, ormaritalcolumns. - Create a new column,
condition_age, based on a calculation of the age in days at which the patient’s condition was diagnosed. - Write the resulting dataset back to Amazon S3 as Snappy-compressed Apache Parquet-format files. No partitions are necessary.
- Given the small resultset, bucket the data such that only one file is written to minimize the impact of too many small files on future query performance.
- Catalog resulting dataset to a new table in the existing AWS Glue Data Catalog.
AWS ELT Options
There are numerous options on AWS to handle the batch transformation use case described above; a non-exhaustive list includes:
- AWS Glue Studio (UI-driven with AWS Glue PySpark Extensions)
- Amazon Glue DataBrew
- Amazon Athena
- Amazon EMR with Apache Spark
- AWS Glue Studio (Apache Spark script)
- AWS Glue Jobs (Legacy jobs)
- Amazon EMR with Presto
- Amazon EMR with Trino
- Amazon EMR with Hive
- AWS Step Functions and AWS Lambda
- Amazon Redshift Spectrum
- Partner solutions on AWS, such as Databricks, Snowflake, Upsolver, StreamSets, Stitch, and Fivetran
- Self-managed custom solutions using a combination of OSS, such as dbt, Airbyte, Dagster, Meltano, Apache NiFi, Apache Drill, Apache Beam, Pandas, Apache Airflow, and Kubernetes
For this comparison, we will choose the first five options listed above to develop our ELT data preparation pipelines: AWS Glue Studio (UI-driven job creation with AWS Glue PySpark Extensions), Amazon Glue DataBrew, Amazon Athena, Amazon EMR with Apache Spark, and AWS Glue Studio (Apache Spark script).

AWS Glue Studio
According to the documentation, “AWS Glue Studio is a new graphical interface that makes it easy to create, run, and monitor extract, transform, and load (ETL) jobs in AWS Glue. You can visually compose data transformation workflows and seamlessly run them on AWS Glue’s Apache Spark-based serverless ETL engine. You can inspect the schema and data results in each step of the job.”
AWS Glue Studio’s visual job creation capability uses the AWS Glue PySpark Extensions, an extension of the PySpark Python dialect for scripting ETL jobs. The extensions provide easier integration with AWS Glue Data Catalog and other AWS-managed data services. As opposed to using the graphical interface for creating jobs with AWS Glue PySpark Extensions, you can also run your Spark scripts with AWS Glue Studio. In fact, we can use the exact same scripts run on Amazon EMR.
For the tests, we are using the G.2X worker type, Glue version 3.0 (Spark 3.1.1 and Python 3.7), and Python as the language choice for this comparison. We will test three worker configurations using both UI-driven job creation with AWS Glue PySpark Extensions and Apache Spark script options:
- 10 workers with a maximum of 20 DPUs
- 20 workers with a maximum of 40 DPUs
- 40 workers with a maximum of 80 DPUs



AWS Glue DataBrew
According to the documentation, “AWS Glue DataBrew is a visual data preparation tool that enables users to clean and normalize data without writing any code. Using DataBrew helps reduce the time it takes to prepare data for analytics and machine learning (ML) by up to 80 percent, compared to custom-developed data preparation. You can choose from over 250 ready-made transformations to automate data preparation tasks, such as filtering anomalies, converting data to standard formats, and correcting invalid values.”
DataBrew allows you to set the maximum number of DataBrew nodes that can be allocated when a job runs. For this comparison, we will test three different node configurations:
- 3 maximum nodes
- 10 maximum nodes
- 20 maximum nodes



Amazon Athena
According to the documentation, “Athena helps you analyze unstructured, semi-structured, and structured data stored in Amazon S3. Examples include CSV, JSON, or columnar data formats such as Apache Parquet and Apache ORC. You can use Athena to run ad-hoc queries using ANSI SQL, without the need to aggregate or load the data into Athena.”
Although Athena is classified as an ad-hoc query engine, using a CREATE TABLE AS SELECT (CTAS) query, we can create a new table in the AWS Glue Data Catalog and write to Amazon S3 from the results of a SELECT statement from another query. That other query statement performs a transformation on the data using SQL.
Amazon Athena is a fully managed AWS service and has no performance settings to adjust or monitor.


CTAS and Partitions
A notable limitation of Amazon Athena for the batch use case is the 100 partition limit with CTAS queries. Athena [only] supports writing to 100 unique partition and bucket combinations with CTAS. Partitioned by year, month, and day, the observations test case requires 2,558 partitions, and the observations test case requires 10,433 partitions. There is a recommended workaround using an INSERT INTO statement. However, the workaround requires additional SQL logic, computation, and most important cost. It is not practical, in my opinion, compared to other methods when a higher number of partitions are needed. To avoid the partition limit with CTAS, we will only partition by year and bucket by month when using Athena. Take this limitation into account when comparing the final results.
Amazon EMR with Apache Spark
According to the documentation, “Amazon EMR is a cloud big data platform for running large-scale distributed data processing jobs, interactive SQL queries, and machine learning (ML) applications using open-source analytics frameworks such as Apache Spark, Apache Hive, and Presto. You can quickly and easily create managed Spark clusters from the AWS Management Console, AWS CLI, or the Amazon EMR API.”
For this comparison, we are using two different Spark 3.1.2 EMR clusters:
- (1) r5.xlarge Master node and (2) r5.2xlarge Core nodes
- (1) r5.2xlarge Master node and (4) r5.2xlarge Core nodes
All Spark jobs are written in both Python (PySpark) and Scala. We are using the AWS Glue Data Catalog as the metastore for Spark SQL instead of Apache Hive.



Results
Data pipelines were developed and tested for each of the three test cases using the five chosen AWS ELT services and configuration variations. Each pipeline was then run 3–5 times, for a total of approximately 150 runs. The resulting AWS Glue Data Catalog table and data in Amazon S3 were deleted between each pipeline run. Each new run created a new data catalog table and wrote new results to Amazon S3. The median execution times from these tests are shown below.


Although we can make some general observations about the execution times of the chosen AWS services, the results are not meant to be a definitive guide to performance. An accurate comparison would require a deeper understanding of how each of these managed services works under the hood, in order to both optimize and balance their compute profiles correctly.
Amazon Athena
The Resultset column contains the final number of records written to Amazon S3 by Athena. The results contain the data pipeline’s median execution time and any additional data points.

AWS Glue Studio (AWS Glue PySpark Extensions)
Tests were run with three different configurations for AWS Glue Studio using the graphical interface for creating jobs with AWS Glue PySpark Extensions. Times for each configuration were nearly identical.

AWS Glue Studio (Apache PySpark script)
As opposed to using the graphical interface for creating jobs with AWS Glue PySpark Extensions, you can also run your Apache Spark scripts with AWS Glue Studio. The tests were run with the same three configurations as above. The execution times compared to the Amazon EMR tests, below, are almost identical.

Amazon EMR with Apache Spark
Tests were run with three different configurations for Amazon EMR with Apache Spark using PySpark. The first set of results is for the 2-node EMR cluster. The second set of results is for the 4-node cluster. The third set of results is for the same 4-node cluster in which the data was not bucketed into a single file within each partition. Compare the execution times and the number of objects against the previous set of results. Too many small files can negatively impact query performance.

It is commonly stated that “Scala is almost ten times faster than Python.” However, with Amazon EMR, jobs written in Python (PySpark) and Scala had similar execution times for all three test cases.

Amazon Glue DataBrew
Tests were run with three different configurations Amazon Glue DataBrew, including 3, 10, and 20 maximum nodes. Times for each configuration were nearly identical.

Observations
- All tested AWS services can read and write to an AWS Glue Data Catalog and the underlying datastore, Amazon S3. In addition, they all work with the most common analytics data file formats.
- All tested AWS services have rich APIs providing access through the AWS CLI and SDKs, which support multiple programming languages.
- Overall, AWS Glue Studio, using the AWS Glue PySpark Extensions, appears to be the most capable ELT tool of the five services tested and with the best performance.
- Both AWS Glue DataBrew and AWS Glue Studio are no-code or low-code services, democratizing access to data for non-programmers. Conversely, Amazon Athena requires knowledge of ANSI SQL, and Amazon EMR with Apache Spark requires knowledge of Scala or Python. Be cognizant of the potential trade-offs from using no-code or low-code services on observability, configuration control, and automation.
- Both AWS Glue DataBrew and AWS Glue Studio can write a custom Parquet writer type optimized for Dynamic Frames, GlueParquet. One potential advantage, a pre-computed schema is not required before writing.
- There is a slight ‘cold-start’ with Glue Studio. Studio startup times ranged from 7 seconds to 2 minutes and 4 seconds in the tests. However, the lower execution time of AWS Glue Studio compared to Amazon EMR with Spark and AWS Glue DataBrew in the tests offsets any initial cold-start time, in my opinion.
- Changing the maximum number of units from 3 to 10 to 20 for AWS Glue DataBrew made negligible differences in job execution times. Given the nearly identical execution times, it is unclear exactly how many units are being used by the job. More importantly, how many DataBrew node hours we are being billed for. These are some of the trade-offs with a fully-managed service — visibility and fine-tuning configuration.
- Similarly, with AWS Glue Studio, using either 10 workers w/ max. 20 DPUs, 20 workers w/ max. 40 DPUs, or 40 workers w/ max. 80 DPUs resulted in nearly identical executions times.
- Amazon Athena had the fastest execution times but is limited by the 100 partition limit for large CTAS resultsets. Athena is not practical, in my opinion, compared to other ELT methods, when a higher number of partitions are needed.
- It is commonly stated that “Scala is almost ten times faster than Python.” However, with Amazon EMR, jobs written in Python (PySpark) and Scala had almost identical execution times for all three test cases.
- Using Amazon EMR with EC2 instances takes about 9 minutes to provision a new cluster for this comparison fully. Given nearly identical execution times to AWS Glue Studio with Apache Spark scripts, Glue has the clear advantage of nearly instantaneous startup times.
- AWS recently announced Amazon EMR Serverless. Although this service is still in Preview, this new version of EMR could potentially reduce or eliminate the lengthy startup time for ephemeral clusters requirements.
- Although not discussed, scheduling the data pipelines to run each night was a requirement for our use case. AWS Glue Studio jobs and AWS Glue DataBrew jobs are schedulable from those services. For Amazon EMR and Amazon Athena, we could use Amazon Managed Workflows for Apache Airflow (MWAA), AWS Data Pipeline, or AWS Step Functions combined with Amazon CloudWatch Events Rules to schedule the data pipelines.
Conclusion
Customers have many options for ELT — the cleansing, deduplication, refinement, and normalization of raw data. We examined chosen services on AWS, each capable of handling the analytics use case presented. The best choice of tools depends on your specific ELT use case and performance requirements.
This blog represents my own viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners.
Video Demonstration: Lakehouse Automation on AWS with Apache Airflow
Posted by Gary A. Stafford in Analytics, AWS, Build Automation, Cloud, DevOps, Python, SQL, Technology Consulting on December 2, 2021
Programmatically load and upload data from Amazon Redshift to an Amazon S3-based Data Lake using Apache Airflow
Introduction
In the following video demonstration, we will learn how to programmatically load and upload data from Amazon Redshift to an Amazon S3-based Data Lake using Apache Airflow. Since we are on AWS, we will be using the fully-managed Amazon Managed Workflows for Apache Airflow (Amazon MWAA). Using Airflow, we will COPY raw data into staging tables, then merge that staging data into a series of tables. We will then load incremental data into Redshift on a regular schedule. Next, we will join and aggregate data from several tables and UNLOAD the resulting dataset to an Amazon S3-based data lake. Lastly, we will catalog the data in S3 using AWS Glue and query with Amazon Athena.

Demonstration
Source Code
The source code for this demonstration, including the Airflow DAGs, SQL statements, and data files, is open-sourced and located on GitHub.
DAGs
The DAGs included in the GitHub project are:
- redshift_demo__01_create_tables.py
- redshift_demo__02_initial_load.py
- redshift_demo__03_incremental_load.py
- redshift_demo__04_unload_data.py
- redshift_demo__05_catalog_and_query.py
- redshift_demo__06_run_dags_01_to_05.py
- redshift_demo__06B_run_dags_01_to_05.py (alt. ver. w/external notifications module)

This blog represents my own viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners.
Video Demonstration: Building a Data Lake with Apache Airflow
Posted by Gary A. Stafford in Analytics, AWS, Big Data, Build Automation, Cloud, Python on November 12, 2021
Build a simple Data Lake on AWS using a combination of services, including Amazon Managed Workflows for Apache Airflow (Amazon MWAA), AWS Glue, AWS Glue Studio, Amazon Athena, and Amazon S3
Introduction
In the following video demonstration, we will build a simple data lake on AWS using a combination of services, including Amazon Managed Workflows for Apache Airflow (Amazon MWAA), AWS Glue Data Catalog, AWS Glue Crawlers, AWS Glue Jobs, AWS Glue Studio, Amazon Athena, Amazon Relational Database Service (Amazon RDS), and Amazon S3.
Using a series of Airflow DAGs (Directed Acyclic Graphs), we will catalog and move data from three separate data sources into our Amazon S3-based data lake. Once in the data lake, we will perform ETL (or more accurately ELT) on the raw data — cleansing, augmenting, and preparing it for data analytics. Finally, we will perform aggregations on the refined data and write those final datasets back to our data lake. The data lake will be organized around the data lake pattern of bronze (aka raw), silver (aka refined), and gold (aka aggregated) data, popularized by Databricks.

Demonstration
Source Code
The source code for this demonstration, including the Airflow DAGs, SQL files, and data files, is open-sourced and located on GitHub.
DAGs
The DAGs shown in the video demonstration have been renamed for easier project management within the Airflow UI. The DAGs included in the GitHub project are as follows:
- data_lake__01_clean_and_prep_demo.py
- data_lake__02_run_glue_crawlers_source.py
- data_lake__03_run_glue_jobs_raw.py
- data_lake__04_run_glue_jobs_refined.py
- data_lake__05_submit_athena_queries_agg.py
- data_lake__06_run_dags_01_to_05.py
This blog represents my own viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners.
Video Demonstration: Building a Data Lake on AWS
Build a simple Data Lake on AWS using a combination of services, including AWS Glue, AWS Glue Studio, Amazon Athena, and Amazon S3
Introduction
In the following video demonstration, we will build a simple data lake on AWS using a combination of services, including AWS Glue Data Catalog, AWS Glue Crawlers, AWS Glue Jobs, AWS Glue Studio, Amazon Athena, Amazon Relational Database Service (Amazon RDS), and Amazon S3.
We will catalog and move data from three separate data sources into our Amazon S3-based data lake. Once in the data lake, we will perform ETL (or more accurately ELT) on the raw data — cleansing, augmenting, and preparing it for data analytics. Finally, we will perform aggregations on the refined data and write those final datasets back to our data lake. The data lake will be organized around the data lake pattern of bronze (aka raw), silver (aka refined), and gold (aka aggregated) data, popularized by Databricks.

Demonstration
Source Code
The source code for this demonstration, including the SQL statements, is open-sourced and located on GitHub.
This blog represents my own viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners.
Video Demonstration: Ahana Cloud for Presto on AWS using Apache Hive and AWS Glue
Using Ahana Cloud for Presto to perform analytics on AWS using both Apache Hive and AWS Glue as metastores
Introduction
The following series of five videos are an extended version of the demonstration featured in the October 2021 webinar, Build an Open Data Lake on AWS with Presto. An on-demand copy of the live webinar is available on Ahana.io, featuring Dipti Borkar (Ahana Co-Founder and CPO) and I.
In the demonstration, we will build a data lake on AWS using a combination of Ahana Cloud for Presto, Apache Hive, Apache Superset, Amazon S3, AWS Glue, and Amazon Athena. We then analyze the data in Apache Superset using Ahana Cloud for Presto.

Demonstration
The demonstration is divided into five YouTube videos (playlist):
Source Code
All source code for this post and the previous posts in this series are open-sourced and located on GitHub. In the webinar and the videos, the Apache Hive and AWS Glue data catalog tables contain an _athena or _presto suffix. For clarity, in the source code, I have changed those to indicate the metastore they are associated with, _hive or _glue, since either set of tables can be queried Presto. Additionally, in the webinar and the videos, the raw data files were uploaded to Amazon S3 in uncompressed CSV format; this is unnecessary. The CTAS SQL statements both expect GZIP-compressed CSV files. To save time and cost, upload the compressed files, as they are, to Amazon S3.
The following files are used in the demonstration:
README.md: Instructions for demoahana_demo_glue_artists.sql: AWS Glue SQL statementsahana_demo_glue_artworks.sql: AWS Glue SQL statementsahana_demo_hive.sql: Apache Hive SQL statementsjoins.sql: Simple SQL join statementsuperset_charts.sql: SQL statements for Superset chartsmoma_public_artists.txt.gz: Compressed raw artists datamoma_public_artworks.txt.gz: Compressed raw artworks data
This blog represents my own viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners.
IoT Data Analytics at the Edge: Exploring the convergence of IoT, Data Analytics, and Edge Computing with Grafana, Mosquitto, and TimescaleDB on ARM-based devices
Posted by Gary A. Stafford in Analytics, AWS, Cloud, IoT, Python, Raspberry Pi, Software Development on April 15, 2021
This post is a revised version of an earlier post, featuring major version updates of TimescaleDB (v1.7.4-pg12 to v2.0.0-pg12), Grafana (v7.1.5 to v7.5.2), and Mosquitto (v1.6.12 to v2.0.9). All source code and SQL scripts are revised. Note that TimeScaleDB has a current limitation/bug with Docker on ARM later than v2.0.0.
The Edge
Edge computing is a fast-growing technology trend, which involves pushing compute capabilities to a network’s edge. Wikipedia describes edge computing as a distributed computing paradigm that brings computation and data storage closer to the location needed to improve response times and save bandwidth. The term edge commonly refers to a compute node at the edge of a network (edge device), sitting close to the data source and between that data source and external system such as the Cloud. In his recent post, 3 Advantages (And 1 Disadvantage) Of Edge Computing, well-known futurist Bernard Marr argues reduced bandwidth requirements, reduced latency, and enhanced security and privacy as three primary advantages of edge computing.
David Ricketts, Head of Marketing at Quiss Technology PLC, in his post, Cloud and Edge Computing — The Stats You Need to Know for 2018, estimates that the global edge computing market is expected to reach USD 6.72 billion by 2022 at a compound annual growth rate of a whopping 35.4 percent. Realizing the market potential, many major Cloud providers, edge device manufacturers, and integrators are rapidly expanding their edge compute capabilities. AWS, for example, currently offers more than a dozen services in their edge computing category.
Internet of Things
Edge computing is frequently associated with the Internet of Things (IoT). IoT devices, industrial equipment, and sensors generate data transmitted to other internal and external systems, often by way of edge nodes, such as an IoT Gateway. IoT devices typically generate time-series data. According to Wikipedia, a time series is a set of data points indexed in time order — a sequence taken at successive equally spaced points in time. IoT devices typically generate continuous high-volume streams of time-series data, often on a scale of millions of data points per second. IoT data characteristics require IoT platforms to minimally support temporal accuracy, high-volume ingestion and processing, efficient data compression and downsampling, and real-time querying capabilities.
Edge devices such as IoT Gateways, which aggregate and transmit IoT data from these devices to external systems, are generally lower-powered, with limited processors, memory, and storage. Accordingly, IoT platforms must satisfy all the requirements of IoT data while simultaneously supporting resource-constrained environments.
IoT Analytics at the Edge
Leading Cloud providers AWS, Azure, Google Cloud, IBM Cloud, Oracle Cloud, and Alibaba Cloud all offer IoT services. Many offer IoT services with edge computing capabilities. AWS offers AWS IoT Greengrass. Greengrass provides local compute, messaging, data management, sync, and machine learning (ML) inference capabilities to edge devices. Azure offers Azure IoT Edge. Azure IoT Edge provides the ability to run artificial intelligence (AI), Azure and third-party services, and custom business logic on edge devices using standard containers. Google Cloud offers Edge TPU. Edge TPU (Tensor Processing Unit) is Google’s purpose-built application-specific integrated circuit (ASIC), designed to run AI at the edge.
IoT Analytics
Many Cloud providers also offer IoT analytics as part of their suite of IoT services, although not at the edge. AWS offers AWS IoT Analytics, while Azure has Azure Time Series Insights. Google provides IoT analytics, indirectly, through downstream analytic systems and ad hoc analysis using Google BigQuery or advanced analytics and machine learning with Cloud Machine Learning Engine. These services generally all require data to be transmitted to the Cloud for analytics.

The ability to analyze real-time, streaming IoT data at the edge is critical to a rapid feedback loop. IoT edge analytics can accelerate anomaly detection and remediation, improve predictive maintenance capabilities, and expedite proactive inventory replenishment.
IoT Edge Analytics Stack
In my opinion, the ideal IoT edge analytics stack is comprised of lightweight, purpose-built, easily deployable and manageable, platform- and programming language-agnostic, open-source software components. The minimal IoT edge analytics stack should include:
- Lightweight message broker;
- Purpose-built time-series database;
- ANSI-standard SQL ad-hoc query engine;
- Data visualization tool;
- Simple deployment and management framework;
Each component should be purpose-built for IoT.
Lightweight Message Broker
We will use Eclipse Mosquitto as our message broker. According to the project’s description, Mosquitto is an open-source message broker that implements the Message Queuing Telemetry Transport (MQTT) protocol versions 5.0, 3.1.1, and 3.1. Mosquitto is lightweight and suitable for use on all devices, from low-power single-board computers (SBCs) to full-powered servers.
MQTT Client Library
We will interact with Mosquitto using Eclipse Paho. According to the project, the Eclipse Paho project provides open-source, mainly client-side implementations of MQTT and MQTT-SN in a variety of programming languages. MQTT and MQTT for Sensor Networks (MQTT-SN) are light-weight publish/subscribe messaging transports for TCP/IP and connectionless protocols, such as UDP, respectively.
We will be using Paho’s Python Client. The Paho Python Client provides a client class with support for both MQTT v3.1 and v3.1.1 on Python 2.7 or 3.x. The client also provides helper functions to make publishing messages to an MQTT server straightforward.
Time-Series Database
Time-series databases are optimal for storing IoT data. According to InfluxData, makers of a leading time-series database, InfluxDB, a time-series database (TSDB), is a database optimized for time-stamped or time-series data. Time series data are simply measurements or events that are tracked, monitored, downsampled, and aggregated over time. Jiao Xian of Alibaba Cloud has authored an insightful post on the time-series database ecosystem, What Are Time Series Databases? A few leading Cloud providers offer purpose-built time-series databases, though they are not available at the edge. AWS offers Amazon Timestream, and Alibaba Cloud offers Time Series Database.
InfluxDB is an excellent choice for a time-series database. It was my first choice, along with TimescaleDB, when developing this stack. However, InfluxDB Flux’s apparent incompatibilities with some ARM-based architecture ruled it out for inclusion in the stack for this particular post.
We will use TimescaleDB as our time-series database. TimescaleDB is the leading open-source relational database for time-series data. Described as ‘PostgreSQL for time-series,’ TimescaleDB is based on PostgreSQL, which provides full ANSI SQL, rock-solid reliability, and a massive ecosystem. TimescaleDB claims to achieve 10–100x faster queries than PostgreSQL, InfluxDB, and MongoDB, with native optimizations for time-series analytics.
TimescaleDB is designed for performing analytical queries, both through its native support for PostgreSQL’s full range of SQL functionality and additional functions native to TimescaleDB. These time-series optimized functions include Median/Percentile, Cumulative Sum, Moving Average, Increase, Rate, Delta, Time Bucket, Histogram, and Gap Filling.
Ad-hoc Data Query Engine
We have the option of using psql, the terminal-based front-end to PostgreSQL, to execute ad-hoc queries against TimescaleDB. The psql front-end enables you to enter queries interactively, issue them to PostgreSQL, and see the query results.

We also have the option of using pgAdmin, specifically the biarms/pgadmin4 Docker version, to execute ad-hoc queries and perform most other database tasks. pgAdmin is the most popular open-source administration and development platform for PostgreSQL. While several popular Docker versions of pgAdmin only support Linux AMD64 architectures, the biarms/pgadmin4 Docker version supports ARM-based devices.


Data Visualization
For data visualization, we will use Grafana. Grafana allows you to query, visualize, alert on, and understand metrics no matter where they are stored. With Grafana, you can create, explore, and share dashboards, fostering a data-driven culture. Grafana allows you to define thresholds visually and get notified via Slack, PagerDuty, and more. Grafana supports dozens of data sources, including MySQL, PostgreSQL, Elasticsearch, InfluxDB, TimescaleDB, Graphite, Prometheus, Google BigQuery, GraphQL, and Oracle. Grafana is extensible through a large collection of plugins.

Edge Deployment and Management Platform
Docker introduced the current industry standard for containers in 2013. Docker containers are a standardized unit of software that allows developers to isolate apps from their environment. We will use Docker to deploy the IoT edge analytics stack, referred to herein as the GTM Stack, composed of containerized versions of Grafana, TimescaleDB, Eclipse Mosquitto, and pgAdmin, to an ARMv7-based edge node. The acronym, GTM, comes from the three primary OSS projects composing the stack. The abbreviation also suggests Greenwich Mean Time, relating to the precise time-series nature of IoT data.

Running Docker Engine in swarm mode, we can use Docker to deploy the complete IoT edge analytics stack to the swarm, running on the edge node. The deploy command accepts a stack description in the form of a Docker Compose file, a YAML file used to configure the application’s services. With a single command, we can create and start all the services from the configuration file.
Source Code
All source code for this post is available on GitHub. Use the following command to git clone a local copy of the project. Note that the updated version of the source code for this post is in the v2021–03 branch.
git clone --branch v2021-03 --single-branch --depth 1 \
https://github.com/garystafford/iot-analytics-at-the-edge.git
IoT Devices
For this post, I have deployed three Linux ARM-based IoT devices, each connected to a sensor array. Each sensor array contains multiple analog and digital sensors. The sensors record temperature, humidity, air quality (liquefied petroleum gas (LPG), carbon monoxide (CO), and smoke), light, and motion. For more information on the IoT device and sensor hardware involved, please see my previous post.Getting Started with IoT Analytics on AWS
Analyze environmental sensor data from IoT devices in near real-time with AWS IoT Analyticstowardsdatascience.com
Each ARM-based IoT device runs a small Python3-based script, sensor_data_to_mosquitto.py, shown below.
The IoT devices’ script implements the Eclipse Paho MQTT Python client library. An MQTT message containing simultaneous readings from each sensor is sent to a Mosquitto topic on the edge node at a configurable frequency.
Below are the actual sensor readings sent by the IoT device as an MQTT message to the Mosquitto topic.
IoT Edge Node
For this post, I have deployed a single Linux ARM-based edge node. The three IoT devices containing sensor arrays communicate with the edge node over Wi-Fi. IoT devices could easily use an alternative communication protocol, such as BLE, LoRaWAN, or Ethernet. For more information on BLE and LoRaWAN, please see some of my previous posts:LoRa and LoRaWAN for IoT: Getting Started with LoRa and LoRaWAN Protocols for Low Power, Wide Area Networking of IoT and BLE and GATT for IoT: Getting Started with Bluetooth Low Energy (BLE) and Generic Attribute Profile (GATT) Specification for IoT.
The edge node also runs a small Python3-based script, mosquitto_to_timescaledb.py, shown below.
Like the IoT devices, the edge node’s script implements the Eclipse Paho MQTT Python client library. The script pulls MQTT messages off a Mosquitto topic(s), serializes the message payload to JSON, and writes the payload’s data to the TimescaleDB database. The edge node’s script accepts several arguments, which allow you to configure the necessary Mosquitto and TimescaleDB connection settings.
Why not use Telegraf?
Telegraf is a plugin-driven agent that collects, processes, aggregates, and writes metrics. There is a Telegraf output plugin, the PostgreSQL and TimescaleDB Output Plugin for Telegraf, produced by TimescaleDB. The plugin can replace the need to manage and maintain the above script. However, I chose not to use it because it is not yet an official Telegraf plugin. If the plugin was included in a Telegraf release, I would certainly encourage its use.
Script Management
Both Linux-based IoT devices and edge nodes run systemd system and service manager. To ensure the Python scripts keep running in the case of a system restart, we define a systemd unit. Units are objects that systemd knows how to manage. This is a standardized representation of system resources that can be managed by the suite of daemons and manipulated by the provided utilities. Each script has a systemd unit file. Below, we see the gtm_stack_mosquitto unit file, gtm_stack_mosquitto.service.
The gtm_stack_mosq_to_tmscl unit file, gtm_stack_mosq_to_tmscl.service, is nearly identical.
To install the gtm_stack_mosquitto.service systemd unit file on each IoT device, use the following commands:
Installing the gtm_stack_mosq_to_tmscl.service unit file on the edge node is nearly identical.
Docker Stack
The edge node runs the GTM Docker stack, stack.yml, in a swarm. As discussed earlier, the stack contains four containers: Eclipse Mosquitto, TimescaleDB, Grafana, and pgAdmin. The Mosquitto, TimescaleDB, and Grafana containers have paths within the containers bind-mounted to directories on the edge device. With bind-mounting, the container’s configuration and data will persist if the containers are removed and re-created. The containers are running on an isolated overlay network.
The GTM Docker stack is installed using the following commands on the edge node. We will assume Docker and git are pre-installed on the edge node for this post.
First, we will create several local directories on the edge device, which will be used to bind-mount to the Docker container’s directories. Below, we see the bind-mounted local directories with the eventual container’s contents stored within them.

Next, we copy the custom Mosquitto configuration file, mosquitto.conf, included in the project to the edge device’s correct location.
Lastly, we initialize the Docker swarm and deploy the stack.

docker service ls' command, showing the running GTM Stack containersTimescaleDB Setup
With the GTM stack running, we need to create a single Timescale hypertable, sensor_data, in the TimescaleDB demo_iot database to hold the incoming IoT sensor data. Hypertables, according to TimescaleDB, are designed to be easy to manage and to behave like standard PostgreSQL tables. Hypertables are comprised of many interlinked “chunk” tables. Commands made to the hypertable automatically propagate changes down to all of the chunks belonging to that hypertable.
I suggest using psql to execute the required DDL statements, which will create the hypertable and the proceeding views and database user permissions. All SQL statements are included in the project’s statements.sql file. One way to use psql is to install it on your local workstation, then use psql to connect to the remote edge node. I prefer to instantiate a local PostgreSQL Docker container instance running psql. I then use the local container’s psql client to connect to the edge node’s TimescaleDB database. For example, from my local machine, I run the following docker run command to connect to the edge node’s TimescaleDB database on the edge node, located locally at 192.168.1.12.
Although not as practical, you can also access psql from within the TimescaleDB Docker container, running on the actual edge node, using the following docker exec command.
TimescaleDB Continuous Aggregates
For this post’s demonstration, we will create four TimescaleDB materialized views, which will be queried from a Grafana Dashboard. The materialized views are TimescaleDB Continuous Aggregates. According to Timescale, aggregate queries which touch large swathes of time-series data can take a long time to compute because the system needs to scan large amounts of data on every query execution. To make these queries faster, a continuous aggregate allows materializing the computed aggregates, while also providing means to continuously, and with low overhead, keep them up-to-date as the underlying source data changes.
For example, we generate sensor data every five seconds from the three IoT devices in this post. When visualizing a 24-hour period in Grafana, using continuous aggregates with an interval of one minute, we would reduce the total volume of data queried from 51,840 rows to 4,320 rows, a reduction of over 91%. The larger the time period or the number of IoT devices being analyzed, the more significant these savings will positively impact query performance.
A time_bucket on the time partitioning column of the hypertable is required for all continuous aggregate views. The time_bucket function, in this case, has a bucket width (interval) of 1 minute. The interval is configurable.
To automatically refresh the four materialized views, we will create four corresponding continuous aggregate policies. In this demonstration, the continuous aggregate policies create a refresh window between one week ago and one hour ago, with a refresh interval of one hour.
Advanced Analytic Queries
The ability to perform ad-hoc queries on time-series IoT data is an essential feature of the IoT edge analytics stack. We can use psql, pgAdmin, or even our own IDE to perform ad-hoc queries against the TimescaleDB database on the edge node. Below are examples of typical ad-hoc queries a data analyst might perform on IoT sensor data. These example queries demonstrate TimescaleDB’s advanced analytical capabilities for working with time-series data, including Moving Average, Delta, Time Bucket, and Histogram.
Data Visualization with Grafana
Using the TimescaleDB continuous aggregates we have created, we can quickly build a richly featured dashboard in Grafana. Below we see a typical IoT Dashboard you might build to monitor the post’s IoT sensor data in near real-time. An exported version, dashboard_external_export.json, is included in the GitHub project.


Limiting Grafana’s Access to IoT Data
Following the Grafana recommendation for database user permissions, we create a grafanareader PostgresSQL user, and limit the user’s access to the sensor_data table and the four views we created. Grafana will use this user’s credentials to perform SELECT queries of the TimescaleDB demo_iot database.
Using PostgreSQL in Grafana
Grafana’s documentation includes a comprehensive set of instructions for Using PostgreSQL in Grafana. To connect to the TimescaleDB database from Grafana, we use the PostgreSQL data source plugin.

The data displayed in each Panel in the Grafana Dashboard is based on a SQL query. For example, the Average Temperature Panel might use a query similar to the example below. This particular query also converts Celsius to Fahrenheit. Note the use of Grafana Macros (e.g., $__time(), $__timeFilter()). Macros can be used within a query to simplify syntax and allow for dynamic parts.
SELECT
$__time(bucket),
device_id AS metric,
((avg_temp * 1.9) + 32) AS avg_temp
FROM temperature_humidity_summary_minute
WHERE
$__timeFilter(bucket)
ORDER BY 1,2
Below, we see another example from the Average Humidity Panel. In this particular query, we might choose to limit the humidity data to a valid range of 0%–100%.
SELECT
$__time(bucket),
device_id AS metric,
avg_humidity
FROM temperature_humidity_summary_minute
WHERE
$__timeFilter(bucket)
AND avg_humidity >= 0.0
AND avg_humidity <= 100.0
ORDER BY 1,2
Mobile Friendly
Grafana dashboards are mobile-friendly. Below we see two views of the dashboard, using the Chrome mobile browser on an Apple iPhone.

Grafana Alerts
Grafana allows Alerts to be created based on the Rules you define in each Panel. If data values match the Rule’s conditions, which you pre-define, such as a temperature reading above a certain threshold for a set amount of time, an alert is sent to your choice of destinations. According to the Rule shown below, If the average temperature exceeds 75°F for a period of 5 minutes, an alert is sent.

As demonstrated below, when the laboratory temperature began to exceed 75°F, the alert entered a ‘Pending’ state. If the temperature exceeded 75°F for the pre-determined period of 5 minutes, the alert status changes to ‘Alerting’, and an alert is sent. When the temperature dropped back below 75°F for the pre-determined period of 5 minutes, the alert status changed from ‘Alerting’ to ‘OK’, and a subsequent notification was sent.

There are currently twenty alert notifiers available out-of-the-box with Grafana, including Slack, email, PagerDuty, webhooks, VictorOps, Opsgenie, and Microsoft Teams. We can use Grafana Alerts to notify the proper resources, in near real-time, if an issue is detected based on the data. Below, we see an actual series of high-temperature alerts sent by Grafana to the Slack channel, followed by subsequent notifications as the temperature returned to normal.

Conclusion
This post explored the development of an IoT edge analytics stack comprised of lightweight, purpose-built, easily deployable and manageable platform- and programming language-agnostic, open-source software components. These components included Docker containerized versions of Grafana, TimescaleDB, Eclipse Mosquitto, and pgAdmin, referred to as the GTM Stack. Using the GTM stack, we collected, analyzed, and visualized IoT data without first shipping the data to Cloud or other external systems.
This blog represents my own viewpoints and not of my employer, Amazon Web Services (AWS). All product names, logos, and brands are the property of their respective owners.